Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos
- PMID: 24994903
- PMCID: PMC4115566
- DOI: 10.1073/pnas.1410022111
Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos
Abstract
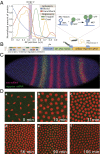
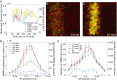
We present the use of recently developed live imaging methods to examine the dynamic regulation of even-skipped (eve) stripe 2 expression in the precellular Drosophila embryo. Nascent transcripts were visualized via MS2 RNA stem loops. The eve stripe 2 transgene exhibits a highly dynamic pattern of de novo transcription, beginning with a broad domain of expression during nuclear cycle 12 (nc12), and progressive refinement during nc13 and nc14. The mature stripe 2 pattern is surprisingly transient, constituting just ∼15 min of the ∼90-min period of expression. Nonetheless, this dynamic transcription profile faithfully predicts the limits of the mature stripe visualized by conventional in situ detection methods. Analysis of individual transcription foci reveals intermittent bursts of de novo transcription, with duration cycles of 4-10 min. We discuss a multistate model of transcription regulation and speculate on its role in the dynamic repression of the eve stripe 2 expression pattern during development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5(5):827–839. - PubMed
-
- Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254(5036):1385–1387. - PubMed
-
- Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122(1):205–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

