Crystal structures of apparent saccharide sensors from histidine kinase receptors prevalent in a human gut symbiont
- PMID: 24995510
- PMCID: PMC4169315
- DOI: 10.1111/febs.12904
Crystal structures of apparent saccharide sensors from histidine kinase receptors prevalent in a human gut symbiont
Abstract
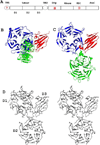
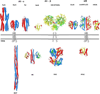
The adult human gut is a complicated ecosystem in which host-bacterium symbiosis plays an important role. Bacteroides thetaiotaomicron is a predominant member of the gut microflora, providing the human digestive tract with a large number of glycolytic enzymes. Expression of many of these enzymes appears to be controlled by histidine kinase receptors that are fused into unusual hybrid two-component systems that share homologous periplasmic sensor domains. These sensor domains belong to the third most populated (HK3) family based on a previous unpublished bioinformatics analysis of predicted histidine kinase sensors. Here, we present the crystal structures of two sensor domains representative of the HK3 family. Each sensor is folded into three domains: two-seven-bladed β-propeller domains and one β-sandwich domain. Both sensors form dimers in crystals, and one sensor appears to be physiologically relevant. The folding characteristics in the individual domains, the domain organization, and the oligomeric architecture are all unique to HK3 sensors. Sequence analysis of the HK3 sensors indicates that these sensor domains are shared among other signaling molecules, implying combinatorial molecular evolution.
Database: The structural data for the crystallographic results for HK3 BT4673S and HK3 BT3049S have been deposited in the Protein Data Bank under accession numbers 3OTT and 3V9F, respectively.
Structured digital abstract: HK3BT3049S and HK3BT3049S bind by x-ray crystallography (View interaction) HK3BT3049S and HK3BT3049S bind by molecular sieving (View interaction) HK3BT3049S and HK3BT3049S bind by cosedimentation through density gradient (View interaction) HK3BT4673s and HK3BT4673s bind by cosedimentation through density gradient (View interaction) HK3BT4673s and HK3BT4673s bind by molecular sieving (View interaction).
Keywords: hybrid two-component system; saccharide sensing; signal transduction; β-propeller domain; β-sandwich domain.
© 2014 FEBS.
Figures





References
-
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. - PubMed
-
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. - PubMed
-
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

