A genome-wide association study of immune response traits in Canadian Holstein cattle
- PMID: 24996426
- PMCID: PMC4099479
- DOI: 10.1186/1471-2164-15-559
A genome-wide association study of immune response traits in Canadian Holstein cattle
Abstract
Background: Breeding for enhanced immune response (IR) has been suggested as a tool to improve inherent animal health. Dairy cows with superior antibody-mediated (AMIR) and cell-mediated immune responses (CMIR) have been demonstrated to have a lower occurrence of many diseases including mastitis. Adaptive immune response traits are heritable, and it is, therefore, possible to breed for improved IR, decreasing the occurrence of disease. The objective of this study was to perform genome-wide association studies to determine differences in genetic profiles among Holstein cows classified as High or Low for AMIR and CMIR. From a total of 680 cows with immune response phenotypes, 163 cows for AMIR (81 High and 82 Low) and 140 for CMIR (75 High and 65 Low) were selectively genotyped using the Illumina Bovine SNP50 BeadChip. Results were validated using an unrelated population of 164 Holstein bulls IR phenotyped for AMIR and 146 for CMIR.
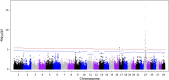
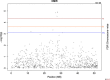
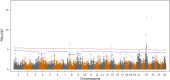
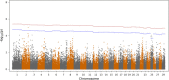
Results: A generalized quasi likelihood score method was used to determine single nucleotide polymorphisms (SNP) and chromosomal regions associated with immune response. After applying a 5% chromosomal false discovery rate, 186 SNPs were significantly associated with AMIR. The majority (93%) of significant markers were on chromosome 23, with a similar peak found in the bull population. For CMIR, 21 SNP markers remained significant. Candidate genes within 250,000 base pairs of significant SNPs were identified to determine biological pathways associated with AMIR and CMIR. Various pathways were identified, including the antigen processing and presentation pathway, important in host defense. Candidate genes included those within the bovine Major Histocompatability Complex such as BoLA-DQ, BoLA-DR and the non-classical BoLA-NC1 for AMIR and BoLA-DQ for CMIR, the complement system including C2 and C4 for AMIR and C1q for CMIR, and cytokines including IL-17A, IL17F for AMIR and IL-17RA for CMIR and tumor necrosis factor for both AMIR and CMIR. Additional genes associated with CMIR included galectins 1, 2 and 3, BCL2 and β-defensin.
Conclusions: The significant genetic variation associated with AMIR and CMIR in this study may imply feasibility to include immune response in genomic breeding indices as an approach to improve inherent animal health.
Figures


Similar articles
-
Genetic parameters of adaptive immune response traits in Canadian Holsteins.J Dairy Sci. 2012 Jan;95(1):401-9. doi: 10.3168/jds.2011-4452. J Dairy Sci. 2012. PMID: 22192219
-
Association of bovine leukocyte antigen (BoLA) DRB3.2 with immune response, mastitis, and production and type traits in Canadian Holsteins.J Dairy Sci. 2007 Feb;90(2):1029-38. doi: 10.3168/jds.S0022-0302(07)71589-8. J Dairy Sci. 2007. PMID: 17235182
-
Phenotypic and genetic parameters of antibody and delayed-type hypersensitivity responses of lactating Holstein cows.Vet Immunol Immunopathol. 2013 Aug 15;154(3-4):83-92. doi: 10.1016/j.vetimm.2013.03.014. Epub 2013 Mar 30. Vet Immunol Immunopathol. 2013. PMID: 23747204
-
Short communication: Cytokine profiles from blood mononuclear cells of dairy cows classified with divergent immune response phenotypes.J Dairy Sci. 2016 Mar;99(3):2364-2371. doi: 10.3168/jds.2015-9449. Epub 2015 Dec 24. J Dairy Sci. 2016. PMID: 26723120
-
Genetic variation and responses to vaccines.Anim Health Res Rev. 2004 Dec;5(2):197-208. doi: 10.1079/ahr200469. Anim Health Res Rev. 2004. PMID: 15984325 Review.
Cited by
-
Cross-Species Meta-Analysis of Transcriptomic Data in Combination With Supervised Machine Learning Models Identifies the Common Gene Signature of Lactation Process.Front Genet. 2018 Jul 12;9:235. doi: 10.3389/fgene.2018.00235. eCollection 2018. Front Genet. 2018. PMID: 30050559 Free PMC article.
-
RNA-Seq Analysis of Peripheral Whole Blood from Dairy Bulls with High and Low Antibody-Mediated Immune Responses-A Preliminary Study.Animals (Basel). 2023 Jul 5;13(13):2208. doi: 10.3390/ani13132208. Animals (Basel). 2023. PMID: 37444006 Free PMC article.
-
Genomic Analysis of IgG Antibody Response to Common Pathogens in Commercial Sows in Health-Challenged Herds.Front Genet. 2020 Oct 23;11:593804. doi: 10.3389/fgene.2020.593804. eCollection 2020. Front Genet. 2020. PMID: 33193739 Free PMC article.
-
Single nucleotide polymorphism mining and nucleotide sequence analysis of Mx1 gene in exonic regions of Japanese quail.Vet World. 2015 Dec;8(12):1435-43. doi: 10.14202/vetworld.2015.1435-1443. Epub 2015 Dec 29. Vet World. 2015. PMID: 27047057 Free PMC article.
-
Genetic and genomic basis of antibody response to porcine reproductive and respiratory syndrome (PRRS) in gilts and sows.Genet Sel Evol. 2016 Jul 14;48(1):51. doi: 10.1186/s12711-016-0230-0. Genet Sel Evol. 2016. PMID: 27417876 Free PMC article.
References
-
- Abdel-Azim GA, Freeman AE, Kehrli ME, Jr, Kelm SC, Burton JL, Kuck AL, Schnell S. Genetic basis and risk factors for infectious and noninfectious diseases in US Holsteins. I. Estimation of genetic parameters for single diseases and general health. J Dairy Sci. 2005;88:1199–1207. doi: 10.3168/jds.S0022-0302(05)72786-7. - DOI - PubMed
-
- Mallard BA, Atalla H, Cartwright S, Hine BC, Hussey B, Paibomesai M, Thompson-Crispi KA, Wagter-Lesperance L. Genetic and Epigenetic Regulation of the Bovine Immune System: Practical Implications of the High Immune Response Technology. 2011. Proc National Mastitis Council 50th Annual Meeting; pp. 53–63.
-
- Wagter L, Mallard BA. Method of Identifying High Immune Response Animals. 2007.
-
- DeLapaz J. Using Humoral and Cellular Response to Novel Antigens in Periparturent Dairy Cows as a Measure of Genetic Disease Resistance in Dairy Cows. 2008. PhD Thesis. MSc Thesis.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

