Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork
- PMID: 24997598
- PMCID: PMC4482249
- DOI: 10.1038/nsmb.2851
Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork
Abstract
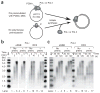
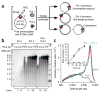
Eukaryotes use distinct polymerases for leading- and lagging-strand replication, but how they target their respective strands is uncertain. We reconstituted Saccharomyces cerevisiae replication forks and found that CMG helicase selects polymerase (Pol) ɛ to the exclusion of Pol δ on the leading strand. Even if Pol δ assembles on the leading strand, Pol ɛ rapidly replaces it. Pol δ-PCNA is distributive with CMG, in contrast to its high stability on primed ssDNA. Hence CMG will not stabilize Pol δ, instead leaving the leading strand accessible for Pol ɛ and stabilizing Pol ɛ. Comparison of Pol ɛ and Pol δ on a lagging-strand model DNA reveals the opposite. Pol δ dominates over excess Pol ɛ on PCNA-primed ssDNA. Thus, PCNA strongly favors Pol δ over Pol ɛ on the lagging strand, but CMG over-rides and flips this balance in favor of Pol ɛ on the leading strand.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
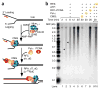
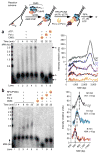
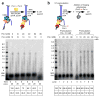
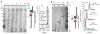
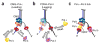
Comment in
-
Delivering nonidentical twins.Nat Struct Mol Biol. 2014 Aug;21(8):649-51. doi: 10.1038/nsmb.2852. Epub 2014 Jul 6. Nat Struct Mol Biol. 2014. PMID: 24997601 Free PMC article.
References
-
- Méndez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. BioEssays. 2003;25:1158–1167. - PubMed
-
- Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. - PubMed
-
- Kornberg A, Baker TA. DNA Replication. W.H. Freeman; New York: 1992.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

