A protein-RNA specificity code enables targeted activation of an endogenous human transcript
- PMID: 24997599
- PMCID: PMC4125476
- DOI: 10.1038/nsmb.2847
A protein-RNA specificity code enables targeted activation of an endogenous human transcript
Abstract
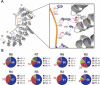
Programmable protein scaffolds that target DNA are invaluable tools for genome engineering and designer control of transcription. RNA manipulation provides broad new opportunities for control, including changes in translation. PUF proteins are an attractive platform for that purpose because they bind specific single-stranded RNA sequences by using short repeated modules, each contributing three amino acids that contact an RNA base. Here, we identified the specificities of natural and designed combinations of those three amino acids, using a large randomized RNA library. The resulting specificity code reveals the RNA binding preferences of natural proteins and enables the design of new specificities. Using the code and a translational activation domain, we designed a protein that targets endogenous cyclin B1 mRNA in human cells, increasing sensitivity to chemotherapeutic drugs. Our study provides a guide for rational design of engineered mRNA control, including translational stimulation.
Figures




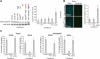
Comment in
-
Expanding the RNA-recognition code of PUF proteins.Nat Struct Mol Biol. 2014 Aug;21(8):653-5. doi: 10.1038/nsmb.2863. Nat Struct Mol Biol. 2014. PMID: 25093524 No abstract available.
-
A protein code to target RNA.Nat Methods. 2014 Sep;11(9):888-9. doi: 10.1038/nmeth.3087. Nat Methods. 2014. PMID: 25317455 No abstract available.
References
-
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18:150–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

