Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification
- PMID: 24997788
- PMCID: PMC4175979
- DOI: 10.1038/nbt.2943
Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification
Abstract
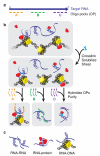
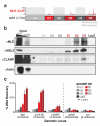
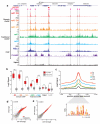
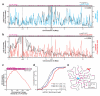
Little is known about the functional domain architecture of long noncoding RNAs (lncRNAs) because of a relative paucity of suitable methods to analyze RNA function at a domain level. Here we describe domain-specific chromatin isolation by RNA purification (dChIRP), a scalable technique to dissect pairwise RNA-RNA, RNA-protein and RNA-chromatin interactions at the level of individual RNA domains in living cells. dChIRP of roX1, a lncRNA essential for Drosophila melanogaster X-chromosome dosage compensation, reveals a 'three-fingered hand' ribonucleoprotein topology. Each RNA finger binds chromatin and the male-specific lethal (MSL) protein complex and can individually rescue male lethality in roX-null flies, thus defining a minimal RNA domain for chromosome-wide dosage compensation. dChIRP improves the RNA genomic localization signal by >20-fold relative to previous techniques, and these binding sites are correlated with chromosome conformation data, indicating that most roX-bound loci cluster in a nuclear territory. These results suggest dChIRP can reveal lncRNA architecture and function with high precision and sensitivity.
Figures

Comment in
-
Non-coding RNA: Zooming in on lncRNA functions.Nat Rev Genet. 2014 Sep;15(9):574-5. doi: 10.1038/nrg3795. Epub 2014 Jul 22. Nat Rev Genet. 2014. PMID: 25048169 No abstract available.
-
Unraveling the lncRNA mystery.Nat Methods. 2014 Sep;11(9):890. doi: 10.1038/nmeth.3089. Nat Methods. 2014. PMID: 25317457 No abstract available.
Similar articles
-
In situ dissection of RNA functional subunits by domain-specific chromatin isolation by RNA purification (dChIRP).Methods Mol Biol. 2015;1262:199-213. doi: 10.1007/978-1-4939-2253-6_12. Methods Mol Biol. 2015. PMID: 25555583
-
Dosage Compensation of the X Chromosome: A Complex Epigenetic Assignment Involving Chromatin Regulators and Long Noncoding RNAs.Annu Rev Biochem. 2018 Jun 20;87:323-350. doi: 10.1146/annurev-biochem-062917-011816. Epub 2018 Apr 18. Annu Rev Biochem. 2018. PMID: 29668306 Review.
-
Chromatin isolation by RNA purification (ChIRP).J Vis Exp. 2012 Mar 25;(61):3912. doi: 10.3791/3912. J Vis Exp. 2012. PMID: 22472705 Free PMC article.
-
RNA-on-X 1 and 2 in Drosophila melanogaster fulfill separate functions in dosage compensation.PLoS Genet. 2018 Dec 10;14(12):e1007842. doi: 10.1371/journal.pgen.1007842. eCollection 2018 Dec. PLoS Genet. 2018. PMID: 30532158 Free PMC article.
-
Identification of the Drosophila X chromosome: The long and short of it.RNA Biol. 2015;12(10):1088-93. doi: 10.1080/15476286.2015.1086864. Epub 2015 Sep 14. RNA Biol. 2015. PMID: 26367502 Free PMC article. Review.
Cited by
-
Long non-coding RNA and chromatin remodeling.RNA Biol. 2015;12(10):1094-8. doi: 10.1080/15476286.2015.1063770. Epub 2015 Jul 15. RNA Biol. 2015. PMID: 26177256 Free PMC article. Review.
-
Differential Expression Pattern of Exosome Long Non-Coding RNAs (lncRNAs) and MicroRNAs (miRNAs) in Vascular Endothelial Cells Under Heat Stroke.Med Sci Monit. 2018 Nov 6;24:7965-7974. doi: 10.12659/MSM.909983. Med Sci Monit. 2018. PMID: 30399613 Free PMC article.
-
Biochemical Methods To Investigate lncRNA and the Influence of lncRNA:Protein Complexes on Chromatin.Biochemistry. 2016 Mar 22;55(11):1615-30. doi: 10.1021/acs.biochem.5b01141. Epub 2016 Feb 24. Biochemistry. 2016. PMID: 26859437 Free PMC article. Review.
-
Unique features of long non-coding RNA biogenesis and function.Nat Rev Genet. 2016 Jan;17(1):47-62. doi: 10.1038/nrg.2015.10. Nat Rev Genet. 2016. PMID: 26666209 Review.
-
DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome.Nature. 2017 Apr 6;544(7648):115-119. doi: 10.1038/nature21715. Epub 2017 Mar 29. Nature. 2017. PMID: 28355180
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

