Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort
- PMID: 24998118
- PMCID: PMC4282620
- DOI: 10.1016/j.eururo.2014.05.039
Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort
Abstract
Background: Risk prediction models that incorporate biomarkers and clinicopathologic variables may be used to improve decision making after radical prostatectomy (RP). We compared two previously validated post-RP classifiers-the Cancer of the Prostate Risk Assessment Postsurgical (CAPRA-S) and the Decipher genomic classifier (GC)-to predict prostate cancer-specific mortality (CSM) in a contemporary cohort of RP patients.
Objective: To evaluate the combined prognostic ability of CAPRA-S and GC to predict CSM.
Design, setting, and participants: A cohort of 1010 patients at high risk of recurrence after RP were treated at the Mayo Clinic between 2000 and 2006. High risk was defined by any of the following: preoperative prostate-specific antigen >20 ng/ml, pathologic Gleason score ≥8, or stage pT3b. A case-cohort random sample identified 225 patients (with cases defined as patients who experienced CSM), among whom CAPRA-S and GC could be determined for 185 patients.
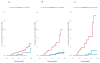
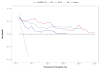
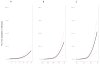
Outcome measurements and statistical analysis: The scores were evaluated individually and in combination using concordance index (c-index), decision curve analysis, reclassification, cumulative incidence, and Cox regression for the prediction of CSM.
Results and limitations: Among 185 men, 28 experienced CSM. The c-indices for CAPRA-S and GC were 0.75 (95% confidence interval [CI], 0.55-0.84) and 0.78 (95% CI, 0.68-0.87), respectively. GC showed higher net benefit on decision curve analysis, but a score combining CAPRA-S and GC did not improve the area under the receiver-operating characteristic curve after optimism-adjusted bootstrapping. In 82 patients stratified to high risk based on CAPRA-S score ≥6, GC scores were likewise high risk for 33 patients, among whom 17 had CSM events. GC reclassified the remaining 49 men as low to intermediate risk; among these men, three CSM events were observed. In multivariable analysis, GC and CAPRA-S as continuous variables were independently prognostic of CSM, with hazard ratios (HRs) of 1.81 (p<0.001 per 0.1-unit change in score) and 1.36 (p=0.01 per 1-unit change in score). When categorized into risk groups, the multivariable HR for high CAPRA-S scores (≥6) was 2.36 (p=0.04) and was 11.26 (p<0.001) for high GC scores (≥0.6). For patients with both high GC and high CAPRA-S scores, the cumulative incidence of CSM was 45% at 10 yr. The study is limited by its retrospective design.
Conclusions: Both GC and CAPRA-S were significant independent predictors of CSM. GC was shown to reclassify many men stratified to high risk based on CAPRA-S ≥6 alone. Patients with both high GC and high CAPRA-S risk scores were at markedly elevated post-RP risk for lethal prostate cancer. If validated prospectively, these findings suggest that integration of a genomic-clinical classifier may enable better identification of those post-RP patients who should be considered for more aggressive secondary therapies and clinical trials.
Patient summary: The Cancer of the Prostate Risk Assessment Postsurgical (CAPRA-S) and the Decipher genomic classifier (GC) were significant independent predictors of prostate cancer-specific mortality. These findings suggest that integration of a genomic-clinical classifier may enable better identification of those post-radical prostatectomy patients who should be considered for more aggressive secondary therapies and clinical trials.
Keywords: Biomarkers; CAPRA-S; Prostate neoplasms; Prostatectomy; RNA; Risk stratification.
Copyright © 2014. Published by Elsevier B.V.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

