A chemical screen identifies small molecules that regulate hepcidin expression
- PMID: 24998898
- PMCID: PMC4198503
- DOI: 10.1016/j.bcmd.2014.06.002
A chemical screen identifies small molecules that regulate hepcidin expression
Abstract
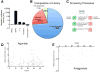
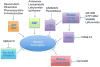
Hepcidin, a peptide hormone produced in the liver, decreases intestinal iron absorption and macrophage iron release via effects on ferroportin. Bone morphogenic protein and Stat3 signaling regulate Hepcidin's transcription. Hepcidin is a potential drug target for patients with iron overload syndromes because its levels are inappropriately low in these individuals. To generate a tool for identifying small molecules that modulate Hepcidin expression, we stably transfected human hepatocytes (HepG2) cells with a reporter construct containing 2.7kb of the human Hepcidin promoter upstream of a firefly reporter gene. We used high throughput methods to screen 10,169 chemicals in duplicate for their effect on Hepcidin expression and cell viability. Regulators were identified as chemicals that caused a change >3 standard deviations above or >1 standard deviation below the mean of the other chemicals (z-score >3 or <1), while not adversely affecting cell viability, quantified by fluorescence assay. Following validation assays, we identified 16 chemicals in a broad range of functional classes that promote Hepcidin expression. All of the chemicals identified increased expression of bone morphogenic protein-dependent and/or Stat3-dependent genes, however none of them strongly increased phosphorylation of Smad1,5,8 or Stat3.
Keywords: Bone morphogenic protein; Hemochromatosis; Interleukin-6; Stat3; Thalassemia.
Copyright © 2014. Published by Elsevier Inc.
Figures
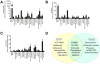
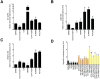

References
-
- Wang RH, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2(6):399–409. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

