Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells
- PMID: 24999991
- PMCID: PMC4085042
- DOI: 10.1371/journal.pone.0099409
Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells
Erratum in
- PLoS One. 2014;9(10):e111296
Abstract
Background: Circulating tumor cells (CTCs) are cancer cells that can be isolated via liquid biopsy from blood and can be phenotypically and genetically characterized to provide critical information for guiding cancer treatment. Current analysis of CTCs is hindered by the throughput, selectivity and specificity of devices or assays used in CTC detection and isolation.
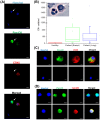
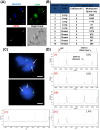
Methodology/principal findings: Here, we enriched and characterized putative CTCs from blood samples of patients with both advanced stage metastatic breast and lung cancers using a novel multiplexed spiral microfluidic chip. This system detected putative CTCs under high sensitivity (100%, n = 56) (Breast cancer samples: 12-1275 CTCs/ml; Lung cancer samples: 10-1535 CTCs/ml) rapidly from clinically relevant blood volumes (7.5 ml under 5 min). Blood samples were completely separated into plasma, CTCs and PBMCs components and each fraction were characterized with immunophenotyping (Pan-cytokeratin/CD45, CD44/CD24, EpCAM), fluorescence in-situ hybridization (FISH) (EML4-ALK) or targeted somatic mutation analysis. We used an ultra-sensitive mass spectrometry based system to highlight the presence of an EGFR-activating mutation in both isolated CTCs and plasma cell-free DNA (cf-DNA), and demonstrate concordance with the original tumor-biopsy samples.
Conclusions/significance: We have clinically validated our multiplexed microfluidic chip for the ultra high-throughput, low-cost and label-free enrichment of CTCs. Retrieved cells were unlabeled and viable, enabling potential propagation and real-time downstream analysis using next generation sequencing (NGS) or proteomic analysis.
Conflict of interest statement
Figures

References
-
- Pantel K, Brakenhoff RH, Brandt B (2008) Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nature Reviews Cancer 8: 329–340. - PubMed
-
- Majid EW, Lim CT (2013) Microfluidic Platforms for Human Disease Cell Mechanics Studies. Materiomics: Multiscale Mechanics of Biological Materials and Structures: Springer. pp. 107–119.
-
- Allan AL, Vantyghem SA, Tuck AB, Chambers AF, Chin-Yee IH, et al. (2005) Detection and quantification of circulating tumor cells in mouse models of human breast cancer using immunomagnetic enrichment and multiparameter flow cytometry. Cytometry Part A 65: 4–14. - PubMed
-
- Gertler R, Rosenberg R, Fuehrer K, Dahm M, Nekarda H, et al. (2003) Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results in Cancer Research 162: 149–156. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

