The evolution of enzyme function in the isomerases
- PMID: 25000289
- PMCID: PMC4139412
- DOI: 10.1016/j.sbi.2014.06.002
The evolution of enzyme function in the isomerases
Abstract
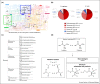
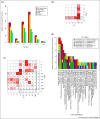
The advent of computational approaches to measure functional similarity between enzymes adds a new dimension to existing evolutionary studies based on sequence and structure. This paper reviews research efforts aiming to understand the evolution of enzyme function in superfamilies, presenting a novel strategy to provide an overview of the evolution of enzymes belonging to an individual EC class, using the isomerases as an exemplar.
Copyright © 2014 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


References
-
- Gerlt J.A., Babbitt P.C. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–246. - PubMed
-
- Todd A.E., Orengo C.A., Thornton J.M. Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol. 2001;307:1113–1143. - PubMed
-
- Khersonsky O., Tawfik D.S. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. - PubMed
-
- Bartlett G.J., Borkakoti N., Thornton J.M. Catalysing new reactions during evolution: economy of residues and mechanism. J Mol Biol. 2003;331:829–860. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

