Identification of two nickel ion-induced genes, NCI16 and PcGST1, in Paramecium caudatum
- PMID: 25001407
- PMCID: PMC4187627
- DOI: 10.1128/EC.00112-14
Identification of two nickel ion-induced genes, NCI16 and PcGST1, in Paramecium caudatum
Abstract
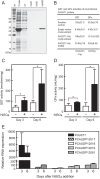
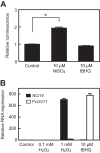
Here, we describe the isolation of two nickel-induced genes in Paramecium caudatum, NCI16 and PcGST1, by subtractive hybridization. NCI16 encoded a predicted four-transmembrane domain protein (∼16 kDa) of unknown function, and PcGST1 encoded glutathione S-transferase (GST; ∼25 kDa) with GST and glutathione peroxidase (GPx) activities. Exposing cells to cobalt chloride also caused the moderate upregulation of NCI16 and PcGST1 mRNAs. Both nickel sulfate and cobalt chloride dose dependently induced NCI16 and PcGST1 mRNAs, but with different profiles. Nickel treatment caused a continuous increase in PcGST1 and NCI16 mRNA levels for up to 3 and 6 days, respectively, and a notable increase in H₂O₂ concentrations in P. caudatum. NCI16 expression was significantly enhanced by incubating cells with H₂O₂, implying that NCI16 induction in the presence of nickel ions is caused by reactive oxygen species (ROS). On the other hand, PcGST1 was highly induced by the antioxidant tert-butylhydroquinone (tBHQ) but not by H2O2, suggesting that different mechanisms mediate the induction of NCI16 and PcGST1. We introduced a luciferase reporter vector with an ∼0.42-kb putative PcGST1 promoter into cells and then exposed the transformants to nickel sulfate. This resulted in significant luciferase upregulation, indicating that the putative PcGST1 promoter contains a nickel-responsive element. Our nickel-inducible system also may be applicable to the efficient expression of proteins that are toxic to host cells or require temporal control.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Sunderman FW., Jr 1989. Mechanisms of nickel carcinogenesis. Scand. J. Work Environ. Health 15:1–12 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

