Optimizing multiplex SNP-based data analysis for genotyping of Mycobacterium tuberculosis isolates
- PMID: 25001491
- PMCID: PMC4117977
- DOI: 10.1186/1471-2164-15-572
Optimizing multiplex SNP-based data analysis for genotyping of Mycobacterium tuberculosis isolates
Abstract
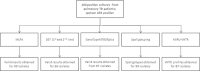
Background: Multiplex ligation-dependent probe amplification (MLPA) is a powerful tool to identify genomic polymorphisms. We have previously developed a single nucleotide polymorphism (SNP) and large sequence polymorphisms (LSP)-based MLPA assay using a read out on a liquid bead array to screen for 47 genetic markers in the Mycobacterium tuberculosis genome. In our assay we obtain information regarding the Mycobacterium tuberculosis lineage and drug resistance simultaneously. Previously we called the presence or absence of a genotypic marker based on a threshold signal level. Here we present a more elaborate data analysis method to standardize and streamline the interpretation of data generated by MLPA. The new data analysis method also identifies intermediate signals in addition to classification of signals as positive and negative. Intermediate calls can be informative with respect to identifying the simultaneous presence of sensitive and resistant alleles or infection with multiple different Mycobacterium tuberculosis strains.
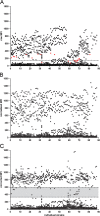
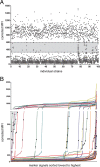
Results: To validate our analysis method 100 DNA isolates of Mycobacterium tuberculosis extracted from cultured patient material collected at the National TB Reference Laboratory of the National Center for Tuberculosis and Lung Diseases in Tbilisi, Republic of Georgia were tested by MLPA. The data generated were interpreted blindly and then compared to results obtained by reference methods. MLPA profiles containing intermediate calls are flagged for expert review whereas the majority of profiles, not containing intermediate calls, were called automatically. No intermediate signals were identified in 74/100 isolates and in the remaining 26 isolates at least one genetic marker produced an intermediate signal.
Conclusion: Based on excellent agreement with the reference methods we conclude that the new data analysis method performed well. The streamlined data processing and standardized data interpretation allows the comparison of the Mycobacterium tuberculosis MLPA results between different experiments. All together this will facilitate the implementation of the MLPA assay in different settings.
Figures
References
-
- Dalla Costa ER, Lazzarini LC, Perizzolo PF, Diaz CA, Spies FS, Costa LL, Ribeiro AW, Barroco C, Schuh SJ, da Silva Pereira MA, Dias CF, Gomes HM, Unis G, Zaha A, da Silva PE A, Suffys PN, Rossetti ML. Mycobacterium tuberculosis of the RDRio genotype is the predominant cause of tuberculosis and associated with multidrug resistance in Porto Alegre City South Brazil. J Clin Microbiol. 2013;51:1071–1077. doi: 10.1128/JCM.01511-12. - DOI - PMC - PubMed
-
- Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, Hilty M, Hopewell PC, Small PM. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

