Whole genome sequence comparison of vtx2-converting phages from Enteroaggregative Haemorrhagic Escherichia coli strains
- PMID: 25001858
- PMCID: PMC4122784
- DOI: 10.1186/1471-2164-15-574
Whole genome sequence comparison of vtx2-converting phages from Enteroaggregative Haemorrhagic Escherichia coli strains
Abstract
Background: Enteroaggregative Haemorrhagic E. coli (EAHEC) is a new pathogenic group of E. coli characterized by the presence of a vtx2-phage integrated in the genomic backbone of Enteroaggregative E. coli (EAggEC). So far, four distinct EAHEC serotypes have been described that caused, beside the large outbreak of infection occurred in Germany in 2011, a small outbreak and six sporadic cases of HUS in the time span 1992-2012. In the present work we determined the whole genome sequence of the vtx2-phage, termed Phi-191, present in the first described EAHEC O111:H2 isolated in France in 1992 and compared it with those of the vtx-phages whose sequences were available.
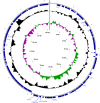
Results: The whole genome sequence of the Phi-191 phage was identical to that of the vtx2-phage P13374 present in the EAHEC O104:H4 strain isolated during the German outbreak 20 years later. Moreover, it was also almost identical to those of the other vtx2-phages of EAHEC O104:H4 strains described so far. Conversely, the Phi-191 phage appeared to be different from the vtx2-phage carried by the EAHEC O111:H21 isolated in the Northern Ireland in 2012.The comparison of the vtx2-phages sequences from EAHEC strains with those from the vtx-phages of typical Verocytotoxin-producing E. coli strains showed the presence of a 900 bp sequence uniquely associated with EAHEC phages and encoding a tail fiber.
Conclusions: At least two different vtx2-phages, both characterized by the presence of a peculiar tail fiber-coding gene, intervened in the emergence of EAHEC. The finding of an identical vtx2-phage in two EAggEC strains isolated after 20 years in spite of the high variability described for vtx-phages is unexpected and suggests that such vtx2-phages are kept under a strong selective pressure.The observation that different EAHEC infections have been traced back to countries where EAggEC infections are endemic and the treatment of human sewage is often ineffective suggests that such countries may represent the cradle for the emergence of the EAHEC pathotype. In these regions, EAggEC of human origin can extensively contaminate the environment where they can meet free vtx-phages likely spread by ruminants excreta.
Figures

Similar articles
-
Spread of a distinct Stx2-encoding phage prototype among Escherichia coli O104:H4 strains from outbreaks in Germany, Norway, and Georgia.J Virol. 2012 Oct;86(19):10444-55. doi: 10.1128/JVI.00986-12. Epub 2012 Jul 18. J Virol. 2012. PMID: 22811533 Free PMC article.
-
Shiga toxin-producing Escherichia coli strains from cattle as a source of the Stx2a bacteriophages present in enteroaggregative Escherichia coli O104:H4 strains.Int J Med Microbiol. 2013 Dec;303(8):595-602. doi: 10.1016/j.ijmm.2013.08.001. Epub 2013 Aug 16. Int J Med Microbiol. 2013. PMID: 24012149
-
Molecular detection of a new pathotype enteroaggregative haemorrhagic Escherichia coli (EAHEC) in Indonesia, 2015.Infect Dis Rep. 2020 Jul 7;12(Suppl 1):8745. doi: 10.4081/idr.2020.8745. eCollection 2020 Jul 7. Infect Dis Rep. 2020. PMID: 32874471 Free PMC article.
-
Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain.Int Microbiol. 2011 Sep;14(3):121-41. doi: 10.2436/20.1501.01.142. Int Microbiol. 2011. PMID: 22101411 Review.
-
Escherichia coli O104:H4 Pathogenesis: an Enteroaggregative E. coli/Shiga Toxin-Producing E. coli Explosive Cocktail of High Virulence.Microbiol Spectr. 2014 Dec;2(6). doi: 10.1128/microbiolspec.EHEC-0008-2013. Microbiol Spectr. 2014. PMID: 26104460 Review.
Cited by
-
Functional and comparative genome analysis of novel virulent actinophages belonging to Streptomyces flavovirens.BMC Microbiol. 2017 Mar 3;17(1):51. doi: 10.1186/s12866-017-0940-7. BMC Microbiol. 2017. PMID: 28257628 Free PMC article.
-
Isolation and Characterization of Novel Bacteriophages to Target Carbapenem-Resistant Acinetobacter baumannii.Antibiotics (Basel). 2024 Jun 29;13(7):610. doi: 10.3390/antibiotics13070610. Antibiotics (Basel). 2024. PMID: 39061292 Free PMC article.
-
Replication Region Analysis Reveals Non-lambdoid Shiga Toxin Converting Bacteriophages.Front Microbiol. 2021 Mar 18;12:640945. doi: 10.3389/fmicb.2021.640945. eCollection 2021. Front Microbiol. 2021. PMID: 33868197 Free PMC article.
-
Escherichia coli O157:H7 strains harbor at least three distinct sequence types of Shiga toxin 2a-converting phages.BMC Genomics. 2015 Sep 29;16:733. doi: 10.1186/s12864-015-1934-1. BMC Genomics. 2015. PMID: 26416807 Free PMC article.
-
Characterization and comparative genomic analysis of virulent and temperate Bacillus megaterium bacteriophages.PeerJ. 2018 Dec 10;6:e5687. doi: 10.7717/peerj.5687. eCollection 2018. PeerJ. 2018. PMID: 30581654 Free PMC article.
References
-
- Paul M, Tsukamoto T, Ghosh AR, Bhattacharya SK, Manna B, Chakrabarti S, Nair GB, Sack DA, Sen D, Takeda Y. The significance of enteroaggregative Escherichia coli in the etiology of hospitalized diarrhoea in Calcutta, India and the demonstration of a new honey-combed pattern of aggregative adherence. FEMS Microbiol Lett. 1994;117(3):319–325. doi: 10.1111/j.1574-6968.1994.tb06786.x. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

