PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER
- PMID: 25002403
- PMCID: PMC4172262
- DOI: 10.1242/jcs.146282
PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER
Abstract
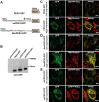
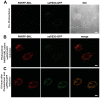
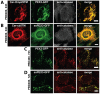
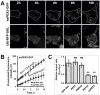
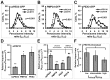
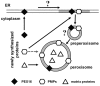
The endoplasmic reticulum (ER) is required for the de novo biogenesis of peroxisomes in mammalian cells. However, its role in peroxisome maintenance is unclear. To explore ER involvement in the maintenance of peroxisomes, we redirect a peroxisomal membrane protein (PMP), PEX3, to directly target to the ER using the N-terminal ER signal sequence from preprolactin. Using biochemical techniques and fluorescent imaging, we find that ER-targeting PEX3 (ssPEX3) is continuously imported into pre-existing peroxisomes. This suggests that the ER constitutively provides membrane proteins and associated lipids to pre-existing peroxisomes. Using quantitative time-lapse live-cell fluorescence microscopy applied to cells that were either depleted of or exogenously expressing PEX16, we find that PEX16 mediates the peroxisomal trafficking of two distinct peroxisomal membrane proteins, PEX3 and PMP34, via the ER. These results not only provide insight into peroxisome maintenance and PMP trafficking in mammalian cells but also highlight important similarities and differences in the mechanisms of PMP import between the mammalian and yeast systems.
Keywords: ER; Live cell imaging; Membrane trafficking; Organelle biogenesis; Peroxisomes; Protein trafficking.
© 2014. Published by The Company of Biologists Ltd.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

