Modulation of frustration in folding by sequence permutation
- PMID: 25002512
- PMCID: PMC4115504
- DOI: 10.1073/pnas.1324230111
Modulation of frustration in folding by sequence permutation
Abstract
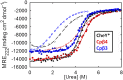
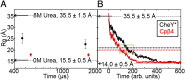
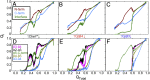
Folding of globular proteins can be envisioned as the contraction of a random coil unfolded state toward the native state on an energy surface rough with local minima trapping frustrated species. These substructures impede productive folding and can serve as nucleation sites for aggregation reactions. However, little is known about the relationship between frustration and its underlying sequence determinants. Chemotaxis response regulator Y (CheY), a 129-amino acid bacterial protein, has been shown previously to populate an off-pathway kinetic trap in the microsecond time range. The frustration has been ascribed to premature docking of the N- and C-terminal subdomains or, alternatively, to the formation of an unproductive local-in-sequence cluster of branched aliphatic side chains, isoleucine, leucine, and valine (ILV). The roles of the subdomains and ILV clusters in frustration were tested by altering the sequence connectivity using circular permutations. Surprisingly, the stability and buried surface area of the intermediate could be increased or decreased depending on the location of the termini. Comparison with the results of small-angle X-ray-scattering experiments and simulations points to the accelerated formation of a more compact, on-pathway species for the more stable intermediate. The effect of chain connectivity in modulating the structures and stabilities of the early kinetic traps in CheY is better understood in terms of the ILV cluster model. However, the subdomain model captures the requirement for an intact N-terminal domain to access the native conformation. Chain entropy and aliphatic-rich sequences play crucial roles in biasing the early events leading to frustration in the folding of CheY.
Keywords: CF-SAXS; CheY permutants; Gō models; protein-folding intermediates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
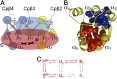
References
-
- Nozaki Y, Schechter NM, Reynolds JA, Tanford C. Use of gel chromatography for the determination of the Stokes radii of proteins in the presence and absence of detergents. A reexamination. Biochemistry. 1976;15(17):3884–3890. - PubMed
-
- Kubelka J, Hofrichter J, Eaton WA. The protein folding ‘speed limit’. Curr Opin Struct Biol. 2004;14(1):76–88. - PubMed
-
- Eaton WA, Thompson PA, Chan CK, Hage SJ, Hofrichter J. Fast events in protein folding. Structure. 1996;4(10):1133–1139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

