Molecular mechanisms for the regulation of histone mRNA stem-loop-binding protein by phosphorylation
- PMID: 25002523
- PMCID: PMC4115514
- DOI: 10.1073/pnas.1406381111
Molecular mechanisms for the regulation of histone mRNA stem-loop-binding protein by phosphorylation
Abstract
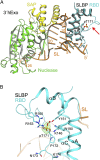
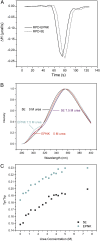
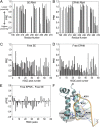
Replication-dependent histone mRNAs end with a conserved stem loop that is recognized by stem-loop-binding protein (SLBP). The minimal RNA-processing domain of SLBP is phosphorylated at an internal threonine, and Drosophila SLBP (dSLBP) also is phosphorylated at four serines in its 18-aa C-terminal tail. We show that phosphorylation of dSLBP increases RNA-binding affinity dramatically, and we use structural and biophysical analyses of dSLBP and a crystal structure of human SLBP phosphorylated on the internal threonine to understand the striking improvement in RNA binding. Together these results suggest that, although the C-terminal tail of dSLBP does not contact the RNA, phosphorylation of the tail promotes SLBP conformations competent for RNA binding and thereby appears to reduce the entropic penalty for the association. Increased negative charge in this C-terminal tail balances positively charged residues, allowing a more compact ensemble of structures in the absence of RNA.
Keywords: NMR; X-ray crystallography; intrinsically disordered protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Marzluff WF, Duronio RJ. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14(6):692–699. - PubMed
-
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA. Gene. 1999;239(1):1–14. - PubMed
-
- Birnstiel ML, Busslinger M, Strub K. Transcription termination and 3′ processing: The end is in site! Cell. 1985;41(2):349–359. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

