Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis
- PMID: 25002590
- PMCID: PMC4156040
- DOI: 10.1074/jbc.M114.579714
Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis
Abstract
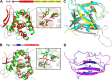
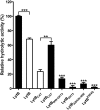
Streptococcus pneumoniae causes a series of devastating infections in humans. Previous studies have shown that the endo-β-N-acetylglucosaminidase LytB is critical for pneumococcal cell division and nasal colonization, but the biochemical mechanism of LytB action remains unknown. Here we report the 1.65 Å crystal structure of the catalytic domain (residues Lys-375-Asp-658) of LytB (termed LytBCAT), excluding the choline binding domain. LytBCAT consists of three structurally independent modules: SH3b, WW, and GH73. These modules form a "T-shaped" pocket that accommodates a putative tetrasaccharide-pentapeptide substrate of peptidoglycan. Structural comparison and simulation revealed that the GH73 module of LytB harbors the active site, including the catalytic residue Glu-564. In vitro assays of hydrolytic activity indicated that LytB prefers the peptidoglycan from the lytB-deficient pneumococci, suggesting the existence of a specific substrate of LytB in the immature peptidoglycan. Combined with in vitro cell-dispersing and in vivo cell separation assays, we demonstrated that all three modules are necessary for the optimal activity of LytB. Further functional analysis showed that the full catalytic activity of LytB is required for pneumococcal adhesion to and invasion into human lung epithelial cells. Structure-based alignment indicated that the unique modular organization of LytB is highly conserved in its orthologs from Streptococcus mitis group and Gemella species. These findings provided structural insights into the pneumococcal cell wall remodeling and novel hints for the rational design of therapeutic agents against pneumococcal growth and thereby the related diseases.
Keywords: Bacterial Pathogenesis; Cell Wall Remodeling; Enzyme Structure; Peptidoglycan; Streptococcus; Structural Biology.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Structural Comparison and Simulation of Pneumococcal Peptidoglycan Hydrolase LytB.Methods Mol Biol. 2016;1440:271-83. doi: 10.1007/978-1-4939-3676-2_19. Methods Mol Biol. 2016. PMID: 27311678
-
Purification and polar localization of pneumococcal LytB, a putative endo-beta-N-acetylglucosaminidase: the chain-dispersing murein hydrolase.J Bacteriol. 2002 Sep;184(18):4988-5000. doi: 10.1128/JB.184.18.4988-5000.2002. J Bacteriol. 2002. PMID: 12193614 Free PMC article.
-
Substrate recognition and catalysis by LytB, a pneumococcal peptidoglycan hydrolase involved in virulence.Sci Rep. 2015 Nov 5;5:16198. doi: 10.1038/srep16198. Sci Rep. 2015. PMID: 26537571 Free PMC article.
-
The pneumococcal cell wall degrading enzymes: a modular design to create new lysins?Microb Drug Resist. 1997 Summer;3(2):199-211. doi: 10.1089/mdr.1997.3.199. Microb Drug Resist. 1997. PMID: 9185148 Review.
-
Recent trends on the molecular biology of pneumococcal capsules, lytic enzymes, and bacteriophage.FEMS Microbiol Rev. 2004 Nov;28(5):553-80. doi: 10.1016/j.femsre.2004.05.002. FEMS Microbiol Rev. 2004. PMID: 15539074 Review.
Cited by
-
Pneumococcal Surface Proteins as Virulence Factors, Immunogens, and Conserved Vaccine Targets.Front Cell Infect Microbiol. 2022 May 12;12:832254. doi: 10.3389/fcimb.2022.832254. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35646747 Free PMC article. Review.
-
Cell Wall Hydrolases in Bacteria: Insight on the Diversity of Cell Wall Amidases, Glycosidases and Peptidases Toward Peptidoglycan.Front Microbiol. 2019 Feb 28;10:331. doi: 10.3389/fmicb.2019.00331. eCollection 2019. Front Microbiol. 2019. PMID: 30873139 Free PMC article. Review.
-
Hemoglobin stimulates vigorous growth of Streptococcus pneumoniae and shapes the pathogen's global transcriptome.Sci Rep. 2020 Sep 16;10(1):15202. doi: 10.1038/s41598-020-71910-1. Sci Rep. 2020. PMID: 32938947 Free PMC article.
-
Identifying genes associated with invasive disease in S. pneumoniae by applying a machine learning approach to whole genome sequence typing data.Sci Rep. 2019 Mar 11;9(1):4049. doi: 10.1038/s41598-019-40346-7. Sci Rep. 2019. PMID: 30858412 Free PMC article.
-
Streptococcus pneumoniae's Virulence and Host Immunity: Aging, Diagnostics, and Prevention.Front Immunol. 2018 Jun 22;9:1366. doi: 10.3389/fimmu.2018.01366. eCollection 2018. Front Immunol. 2018. PMID: 29988379 Free PMC article. Review.
References
-
- Neidhardt F. C., Ingraham J. L., Schaechter M. (1990) Physiology of the Bacterial Cell: A Molecular Approach, pp. 507, American Society for Microbiology, Washington, D. C
-
- Vollmer W., Joris B., Charlier P., Foster S. (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 - PubMed
-
- Musher D. M. (2010) Streptococcus pneumoniae. In Principles and Practice of Infectious Diseases. 7th Ed., pp. 2623–2641, Elsevier Churchill Livingstone, New York
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

