In silico studies of medicinal compounds against hepatitis C capsid protein from north India
- PMID: 25002815
- PMCID: PMC4076477
- DOI: 10.4137/BBI.S15211
In silico studies of medicinal compounds against hepatitis C capsid protein from north India
Abstract
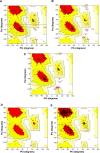
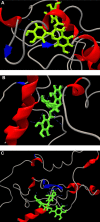
Hepatitis viral infection is a leading cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC). Over one million people are estimated to be persistently infected with hepatitis C virus (HCV) worldwide. As capsid core protein is the key element in spreading HCV; hence, it is considered to be the superlative target of antiviral compounds. Novel drug inhibitors of HCV are in need to complement or replace the current treatments such as pegylated interferon's and ribavirin as they are partially booming and beset with various side effects. Our study was conducted to predict 3D structure of capsid core protein of HCV from northern part of India. Core, the capsid protein of HCV, handles the assembly and packaging of HCV RNA genome and is the least variable of all the ten HCV proteins among the six HCV genotypes. Therefore, we screened four phytochemicals inhibitors that are known to disrupt the interactions of core and other HCV proteins such as (a) epigallocatechin gallate (EGCG), (b) ladanein, (c) naringenin, and (d) silybin extracted from medicinal plants; targeted against active site of residues of HCV-genotype 3 (G3) (Q68867) and its subtypes 3b (Q68861) and 3g (Q68865) from north India. To study the inhibitory activity of the recruited flavonoids, we conducted a quantitative structure-activity relationship (QSAR). Furthermore, docking interaction suggests that EGCG showed a maximum number of hydrogen bond (H-bond) interactions with all the three modeled capsid proteins with high interaction energy followed by naringenin and silybin. Thus, our results strongly correlate the inhibitory activity of the selected bioflavonoid. Finally, the dynamic predicted capsid protein molecule of HCV virion provides a general avenue to target structure-based antiviral compounds that support the hypothesis that the screened inhibitors for viral capsid might constitute new class of potent agents but further confirmation is necessary using in vitro and in vivo studies.
Keywords: capsid protein; docking; hepatitis C virus; hepatocellular carcinoma; inhibitors.
Figures



Similar articles
-
Computational Docking Study of p7 Ion Channel from HCV Genotype 3 and Genotype 4 and Its Interaction with Natural Compounds.PLoS One. 2015 Jun 1;10(6):e0126510. doi: 10.1371/journal.pone.0126510. eCollection 2015. PLoS One. 2015. PMID: 26030803 Free PMC article.
-
Direct binding of a hepatitis C virus inhibitor to the viral capsid protein.PLoS One. 2012;7(2):e32207. doi: 10.1371/journal.pone.0032207. Epub 2012 Feb 28. PLoS One. 2012. PMID: 22389688 Free PMC article.
-
Anti-hepatitis C virus activity and synergistic effect of Nymphaea alba extracts and bioactive constituents in liver infected cells.Microb Pathog. 2018 Aug;121:198-209. doi: 10.1016/j.micpath.2018.05.023. Epub 2018 May 31. Microb Pathog. 2018. PMID: 29775725
-
Does antiviral therapy reduce the risk of hepatocellular carcinoma in patients with chronic hepatitis C?Minerva Gastroenterol Dietol. 2012 Mar;58(1):65-79. Minerva Gastroenterol Dietol. 2012. PMID: 22419005 Review.
-
Challenges in modern drug discovery: a case study of boceprevir, an HCV protease inhibitor for the treatment of hepatitis C virus infection.Acc Chem Res. 2008 Jan;41(1):50-9. doi: 10.1021/ar700109k. Acc Chem Res. 2008. PMID: 18193821 Review.
Cited by
-
Beneficial effects of naringenin in liver diseases: Molecular mechanisms.World J Gastroenterol. 2018 Apr 28;24(16):1679-1707. doi: 10.3748/wjg.v24.i16.1679. World J Gastroenterol. 2018. PMID: 29713125 Free PMC article. Review.
-
Hepatitis C virus and neurological damage.World J Hepatol. 2016 Apr 28;8(12):545-56. doi: 10.4254/wjh.v8.i12.545. World J Hepatol. 2016. PMID: 27134702 Free PMC article. Review.
-
A therapeutic approach for the hepatitis C virus: in silico design of an antisense oligonucleotide-based candidate capsid inhibitor.Virus Genes. 2024 Oct;60(5):446-454. doi: 10.1007/s11262-024-02088-1. Epub 2024 Jul 31. Virus Genes. 2024. PMID: 39083128
-
The potential of epigallocatechin gallate in the chemoprevention and therapy of hepatocellular carcinoma.Front Pharmacol. 2023 May 24;14:1201085. doi: 10.3389/fphar.2023.1201085. eCollection 2023. Front Pharmacol. 2023. PMID: 37292151 Free PMC article. Review.
-
Unlocking Benzosampangine's Potential: A Computational Approach to Investigating, Its Role as a PD-L1 Inhibitor in Tumor Immune Evasion via Molecular Docking, Dynamic Simulation, and ADMET Profiling.Bioinform Biol Insights. 2024 Nov 18;18:11779322241298591. doi: 10.1177/11779322241298591. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 39564188 Free PMC article.
References
-
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–917. - PubMed
-
- Book World Cancer Report 2008 IARC PUBLICATIONS Edited by Peter Boyle and Bernard Levin. 2008. http://www.iarc.fr/en/publications/pdfs-online/wcr/2008/IARC. IARC World Cancer Report.
-
- Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36(1):35–46. - PubMed
-
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5(6):453–63. - PubMed
-
- Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436(7053):933–8. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

