The Ginkgo biloba Extract EGb 761 Modulates Proteasome Activity and Polyglutamine Protein Aggregation
- PMID: 25002904
- PMCID: PMC4068065
- DOI: 10.1155/2014/940186
The Ginkgo biloba Extract EGb 761 Modulates Proteasome Activity and Polyglutamine Protein Aggregation
Abstract
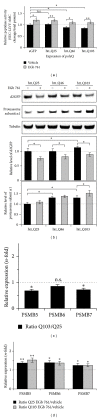
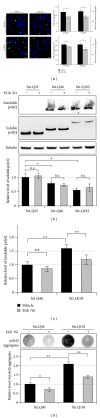
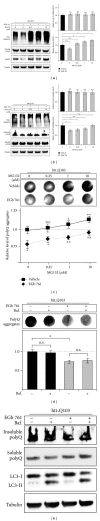
The standardized Ginkgo biloba extract EGb 761 has well-described antioxidative activities and effects on different cytoprotective signaling pathways. Consequently, a potential use of EGb 761 in neurodegenerative diseases has been proposed. A common characteristic feature of a variety of such disorders is the pathologic formation of protein aggregates, suggesting a crucial role for protein homeostasis. In this study, we show that EGb 761 increased the catalytic activity of the proteasome and enhanced protein degradation in cultured cells. We further investigated this effect in a cellular model of Huntington's disease (HD) by employing cells expressing pathologic variants of a polyglutamine protein (polyQ protein). We show that EGb 761 affected these cells by (i) increasing proteasome activity and (ii) inducing a more efficient degradation of aggregation-prone proteins. These results demonstrate a novel activity of EGb 761 on protein aggregates by enhancing proteasomal protein degradation, suggesting a therapeutic use in neurodegenerative disorders with a disturbed protein homeostasis.
Figures

Similar articles
-
EGb 761: ginkgo biloba extract, Ginkor.Drugs R D. 2003;4(3):188-93. doi: 10.2165/00126839-200304030-00009. Drugs R D. 2003. PMID: 12757407 Review.
-
Ginkgo biloba Extract EGb 761 and Its Specific Components Elicit Protective Protein Clearance Through the Autophagy-Lysosomal Pathway in Tau-Transgenic Mice and Cultured Neurons.J Alzheimers Dis. 2018;65(1):243-263. doi: 10.3233/JAD-180426. J Alzheimers Dis. 2018. PMID: 30010136
-
A double-blind, placebo-controlled, randomized trial of Ginkgo biloba extract EGb 761 in a sample of cognitively intact older adults: neuropsychological findings.Hum Psychopharmacol. 2002 Aug;17(6):267-77. doi: 10.1002/hup.412. Hum Psychopharmacol. 2002. PMID: 12404671 Clinical Trial.
-
Treatment with a Ginkgo biloba extract, EGb 761, inhibits excitotoxicity in an animal model of spinocerebellar ataxia type 17.Drug Des Devel Ther. 2016 Feb 18;10:723-31. doi: 10.2147/DDDT.S98156. eCollection 2016. Drug Des Devel Ther. 2016. PMID: 26937174 Free PMC article.
-
Ginkgo biloba extract (EGb 761) in Alzheimer's disease: is there any evidence?Curr Alzheimer Res. 2007 Jul;4(3):253-62. doi: 10.2174/156720507781077304. Curr Alzheimer Res. 2007. PMID: 17627482 Review.
Cited by
-
Modulation of the ubiquitin-proteasome system by marine natural products.Redox Biol. 2021 May;41:101897. doi: 10.1016/j.redox.2021.101897. Epub 2021 Feb 17. Redox Biol. 2021. PMID: 33640701 Free PMC article. Review.
-
Natural Phenolic Compounds with Neuroprotective Effects.Neurochem Res. 2024 Feb;49(2):306-326. doi: 10.1007/s11064-023-04046-z. Epub 2023 Nov 8. Neurochem Res. 2024. PMID: 37940760 Review.
-
The Effect of Polyphenols on Protein Degradation Pathways: Implications for Neuroprotection.Molecules. 2017 Jan 19;22(1):159. doi: 10.3390/molecules22010159. Molecules. 2017. PMID: 28106854 Free PMC article. Review.
-
An Overview of Crucial Dietary Substances and Their Modes of Action for Prevention of Neurodegenerative Diseases.Cells. 2020 Feb 28;9(3):576. doi: 10.3390/cells9030576. Cells. 2020. PMID: 32121302 Free PMC article. Review.
-
Pueraria lobata and Daidzein Reduce Cytotoxicity by Enhancing Ubiquitin-Proteasome System Function in SCA3-iPSC-Derived Neurons.Oxid Med Cell Longev. 2019 Oct 7;2019:8130481. doi: 10.1155/2019/8130481. eCollection 2019. Oxid Med Cell Longev. 2019. PMID: 31687087 Free PMC article.
References
-
- van Beek TA. Chemical analysis of Ginkgo biloba leaves and extracts. Journal of Chromatography A. 2002;967(1):21–55. - PubMed
-
- Mahadevan S, Park Y. Multifaceted therapeutic benefits of Ginkgo biloba L.: chemistry, efficacy, safety, and uses. Journal of Food Science. 2008;73(1):R14–R19. - PubMed
-
- Chan P-C, Xia Q, Fu PP. Ginkgo biloba leave extract: biological, medicinal, and toxicological effects. Journal of Environmental Science and Health C: Environmental Carcinogenesis & Ecotoxicology Reviews. 2007;25:211–244. - PubMed
-
- Janßen IM, Sturtz S, Skipka G, Zentner A, Garrido MV, Busse R. Ginkgo biloba in Alzheimer’s disease: a systematic review. Wiener Medizinische Wochenschrift. 2010;160(21-22):539–546. - PubMed
-
- Horáková L, Licht A, Sandig G, Jakstadt M, Duracková Z, Grune T. Standardized extracts of flavonoids increase the viability of PC12 cells treated with hydrogen peroxide: effects on oxidative injury. Archives of Toxicology. 2003;77(1):22–29. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

