Applying compressed sensing to genome-wide association studies
- PMID: 25002967
- PMCID: PMC4078394
- DOI: 10.1186/2047-217X-3-10
Applying compressed sensing to genome-wide association studies
Abstract
Background: The aim of a genome-wide association study (GWAS) is to isolate DNA markers for variants affecting phenotypes of interest. This is constrained by the fact that the number of markers often far exceeds the number of samples. Compressed sensing (CS) is a body of theory regarding signal recovery when the number of predictor variables (i.e., genotyped markers) exceeds the sample size. Its applicability to GWAS has not been investigated.
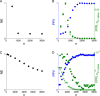
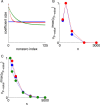
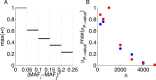
Results: Using CS theory, we show that all markers with nonzero coefficients can be identified (selected) using an efficient algorithm, provided that they are sufficiently few in number (sparse) relative to sample size. For heritability equal to one (h (2) = 1), there is a sharp phase transition from poor performance to complete selection as the sample size is increased. For heritability below one, complete selection still occurs, but the transition is smoothed. We find for h (2) ∼ 0.5 that a sample size of approximately thirty times the number of markers with nonzero coefficients is sufficient for full selection. This boundary is only weakly dependent on the number of genotyped markers.
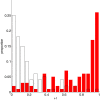
Conclusion: Practical measures of signal recovery are robust to linkage disequilibrium between a true causal variant and markers residing in the same genomic region. Given a limited sample size, it is possible to discover a phase transition by increasing the penalization; in this case a subset of the support may be recovered. Applying this approach to the GWAS analysis of height, we show that 70-100% of the selected markers are strongly correlated with height-associated markers identified by the GIANT Consortium.
Keywords: Compressed sensing; GWAS; Genomic selection; Lasso; Phase transition; Sparsity; Underdetermined system.
Figures






References
-
- Goddard ME, Wray NR, Verbyla K, Visscher PM. Estimating effects and making predictions from genome-wide marker data. Stat Sci. 2009;24:517–529. doi: 10.1214/09-STS306. - DOI
-
- Genovese CR, Jin J, Wasserman L, Yao Z. A comparison of the lasso and marginal regression. J Mach Learn Res. 2012;13:2107–2143.
LinkOut - more resources
Full Text Sources
Other Literature Sources

