JG6, a novel marine-derived oligosaccharide, suppresses breast cancer metastasis via binding to cofilin
- PMID: 25003327
- PMCID: PMC4116503
- DOI: 10.18632/oncotarget.1959
JG6, a novel marine-derived oligosaccharide, suppresses breast cancer metastasis via binding to cofilin
Abstract
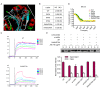
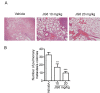
Cofilin, an actin-binding protein which disassembles actin filaments, plays an important role in invasion and metastasis. Here, we discover that JG6, an oligomannurarate sulfate, binds to cofilin, suppresses the migration of human breast cancer cells and cancer metastasis in breast cancer xenograft model. Mechanistically, JG6 occupies actin-binding sites of cofilin, thereby disrupting cofilin modulated actin turnover. Our results highlight the significance of cofilin in cancer and suggest JG6, a cofilin inhibitor, to treat metastatic cancer.
Figures



Similar articles
-
The mitotic kinase Aurora-A induces mammary cell migration and breast cancer metastasis by activating the Cofilin-F-actin pathway.Cancer Res. 2010 Nov 15;70(22):9118-28. doi: 10.1158/0008-5472.CAN-10-1246. Epub 2010 Nov 2. Cancer Res. 2010. PMID: 21045147
-
Memo is a cofilin-interacting protein that influences PLCgamma1 and cofilin activities, and is essential for maintaining directionality during ErbB2-induced tumor-cell migration.J Cell Sci. 2009 Mar 15;122(Pt 6):787-97. doi: 10.1242/jcs.032094. Epub 2009 Feb 17. J Cell Sci. 2009. PMID: 19223396
-
Cytochalasin D acts as an inhibitor of the actin-cofilin interaction.Biochem Biophys Res Commun. 2012 Jul 20;424(1):52-7. doi: 10.1016/j.bbrc.2012.06.063. Epub 2012 Jun 20. Biochem Biophys Res Commun. 2012. PMID: 22728040
-
The cofilin activity cycle in lamellipodia and invadopodia.J Cell Biochem. 2009 Dec 15;108(6):1252-62. doi: 10.1002/jcb.22372. J Cell Biochem. 2009. PMID: 19862699 Free PMC article. Review.
-
Ins and outs of ADF/cofilin activity and regulation.Eur J Cell Biol. 2008 Sep;87(8-9):649-67. doi: 10.1016/j.ejcb.2008.04.001. Epub 2008 May 21. Eur J Cell Biol. 2008. PMID: 18499298 Review.
Cited by
-
Targeting the cytoskeleton against metastatic dissemination.Cancer Metastasis Rev. 2021 Mar;40(1):89-140. doi: 10.1007/s10555-020-09936-0. Epub 2021 Jan 20. Cancer Metastasis Rev. 2021. PMID: 33471283 Review.
-
Overexpression of cofilin correlates with poor survival in breast cancer: A tissue microarray analysis.Oncol Lett. 2017 Aug;14(2):2288-2294. doi: 10.3892/ol.2017.6413. Epub 2017 Jun 19. Oncol Lett. 2017. PMID: 28781665 Free PMC article.
-
Degradation of cofilin is regulated by Cbl, AIP4 and Syk resulting in increased migration of LMP2A positive nasopharyngeal carcinoma cells.Sci Rep. 2017 Aug 21;7(1):9012. doi: 10.1038/s41598-017-09540-3. Sci Rep. 2017. PMID: 28827787 Free PMC article.
-
A novel and selective inhibitor of PKC ζ potently inhibits human breast cancer metastasis in vitro and in mice.Tumour Biol. 2016 Jun;37(6):8391-401. doi: 10.1007/s13277-015-4744-9. Epub 2016 Jan 5. Tumour Biol. 2016. PMID: 26733166
-
Unique cellular protrusions mediate breast cancer cell migration by tethering to osteogenic cells.NPJ Breast Cancer. 2020 Sep 7;6:42. doi: 10.1038/s41523-020-00183-8. eCollection 2020. NPJ Breast Cancer. 2020. PMID: 32964116 Free PMC article.
References
-
- Limame R, Op de Beeck K, Van Laere S, Croes L, De Wilde A, Dirix L, Van Camp G, Peeters M, De Wever O, Lardon F, Pauwels P. Expression profiling of migrated and invaded breast cancer cells predicts early metastatic relapse and reveals Kruppel-like factor 9 as a potential suppressor of invasive growth in breast cancer. Oncoscience. 2014;1:69–81. - PMC - PubMed
-
- Bugyi B, Carlier MF. Control of actin filament treadmilling in cell motility. Annu Rev Biophys. 2010;39:449–470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

