Tipping elements in the human intestinal ecosystem
- PMID: 25003530
- PMCID: PMC4102116
- DOI: 10.1038/ncomms5344
Tipping elements in the human intestinal ecosystem
Abstract
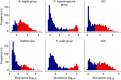
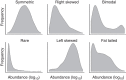
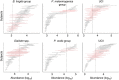
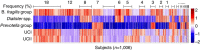
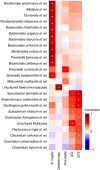
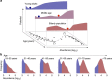
The microbial communities living in the human intestine can have profound impact on our well-being and health. However, we have limited understanding of the mechanisms that control this complex ecosystem. Here, based on a deep phylogenetic analysis of the intestinal microbiota in a thousand western adults, we identify groups of bacteria that exhibit robust bistable abundance distributions. These bacteria are either abundant or nearly absent in most individuals, and exhibit decreased temporal stability at the intermediate abundance range. The abundances of these bimodally distributed bacteria vary independently, and their abundance distributions are not affected by short-term dietary interventions. However, their contrasting alternative states are associated with host factors such as ageing and overweight. We propose that the bistable groups reflect tipping elements of the intestinal microbiota, whose critical transitions may have profound health implications and diagnostic potential.
Figures


References
-
- Zoetendal E. G., Rajilić-Stojanović M. & de Vos W. M. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57, 1605–1615 (2008). - PubMed
-
- de Vos W. M. & de Vos E. A. J. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr. Rev. 70, S45–S56 (2012). - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

