HIV-1 pol diversity among female bar and hotel workers in Northern Tanzania
- PMID: 25003939
- PMCID: PMC4087014
- DOI: 10.1371/journal.pone.0102258
HIV-1 pol diversity among female bar and hotel workers in Northern Tanzania
Abstract
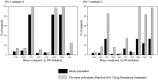
A national ART program was launched in Tanzania in October 2004. Due to the existence of multiple HIV-1 subtypes and recombinant viruses co-circulating in Tanzania, it is important to monitor rates of drug resistance. The present study determined the prevalence of HIV-1 drug resistance mutations among ART-naive female bar and hotel workers, a high-risk population for HIV-1 infection in Moshi, Tanzania. A partial HIV-1 pol gene was analyzed by single-genome amplification and sequencing in 45 subjects (622 pol sequences total; median number of sequences per subject, 13; IQR 5-20) in samples collected in 2005. The prevalence of HIV-1 subtypes A1, C, and D, and inter-subtype recombinant viruses, was 36%, 29%, 9% and 27%, respectively. Thirteen different recombination patterns included D/A1/D, C/A1, A1/C/A1, A1/U/A1, C/U/A1, C/A1, U/D/U, D/A1/D, A1/C, A1/C, A2/C/A2, CRF10_CD/C/CRF10_CD and CRF35_AD/A1/CRF35_AD. CRF35_AD was identified in Tanzania for the first time. All recombinant viruses in this study were unique, suggesting ongoing recombination processes among circulating HIV-1 variants. The prevalence of multiple infections in this population was 16% (n = 7). Primary HIV-1 drug resistance mutations to RT inhibitors were identified in three (7%) subjects (K65R plus Y181C; N60D; and V106M). In some subjects, polymorphisms were observed at the RT positions 41, 69, 75, 98, 101, 179, 190, and 215. Secondary mutations associated with NNRTIs were observed at the RT positions 90 (7%) and 138 (6%). In the protease gene, three subjects (7%) had M46I/L mutations. All subjects in this study had HIV-1 subtype-specific natural polymorphisms at positions 36, 69, 89 and 93 that are associated with drug resistance in HIV-1 subtype B. These results suggested that HIV-1 drug resistance mutations and natural polymorphisms existed in this population before the initiation of the national ART program. With increasing use of ARV, these results highlight the importance of drug resistance monitoring in Tanzania.
Conflict of interest statement
Figures


Similar articles
-
Genetic characterization of HIV-1 strains in Togo reveals a high genetic complexity and genotypic drug-resistance mutations in ARV naive patients.Infect Genet Evol. 2009 Jul;9(4):646-52. doi: 10.1016/j.meegid.2009.04.002. Epub 2009 Apr 15. Infect Genet Evol. 2009. PMID: 19460333
-
HIV-1 subtypes and recombinants in Northern Tanzania: distribution of viral quasispecies.PLoS One. 2012;7(10):e47605. doi: 10.1371/journal.pone.0047605. Epub 2012 Oct 31. PLoS One. 2012. PMID: 23118882 Free PMC article.
-
Surveillance of HIV-1 pol transmitted drug resistance in acutely and recently infected antiretroviral drug-naïve persons in rural western Kenya.PLoS One. 2017 Feb 8;12(2):e0171124. doi: 10.1371/journal.pone.0171124. eCollection 2017. PLoS One. 2017. PMID: 28178281 Free PMC article.
-
Genetic characterization of HIV type 1 among patients with suspected immune reconstitution inflammatory syndrome after initiation of antiretroviral therapy in Kenya.AIDS Res Hum Retroviruses. 2010 Jul;26(7):833-8. doi: 10.1089/aid.2009.0229. AIDS Res Hum Retroviruses. 2010. PMID: 20624074
-
Evaluating HIV drug resistance in the middle East and North Africa and its associated factors: a systematic review.Virol J. 2025 Apr 22;22(1):112. doi: 10.1186/s12985-025-02740-8. Virol J. 2025. PMID: 40264168 Free PMC article.
Cited by
-
The Transmission and Evolution of HIV-1 Quasispecies within One Couple: a Follow-up Study based on Next-Generation Sequencing.Sci Rep. 2018 Jan 23;8(1):1404. doi: 10.1038/s41598-018-19783-3. Sci Rep. 2018. PMID: 29362487 Free PMC article.
-
Low Incidence of HIV-1C Acquired Drug Resistance 10 Years after Roll-Out of Antiretroviral Therapy in Ethiopia: A Prospective Cohort Study.PLoS One. 2015 Oct 29;10(10):e0141318. doi: 10.1371/journal.pone.0141318. eCollection 2015. PLoS One. 2015. PMID: 26512902 Free PMC article.
References
-
- Palella FJ Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, et al. (1998) Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med 338: 853–860. - PubMed
-
- Whitman S, Murphy J, Cohen M, Sherer R (2000) Marked declines in human immunodeficiency virus-related mortality in Chicago in women, African Americans, Hispanics, young adults, and injection drug users, from 1995 through 1997. Arch Intern Med 160: 365–369. - PubMed
-
- Bangsberg DR, Perry S, Charlebois ED, Clark RA, Roberston M, et al. (2001) Non-adherence to highly active antiretroviral therapy predicts progression to AIDS. AIDS 15: 1181–1183. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

