A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy
- PMID: 25004100
- PMCID: PMC4121513
- DOI: 10.1038/mtna.2014.23
A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy
Abstract
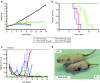
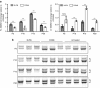
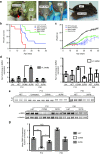
Recent reports underscore the unparalleled potential of antisense-oligonucleotide (ASO)-based approaches to ameliorate various pathological conditions. However, in vivo studies validating the effectiveness of a short ASO (<10-mer) in the context of a human disease have not been performed. One disease with proven amenability to ASO-based therapy is spinal muscular atrophy (SMA). SMA is a neuromuscular disease caused by loss-of-function mutations in the survival motor neuron 1 (SMN1) gene. Correction of aberrant splicing of the remaining paralog, SMN2, can rescue mouse models of SMA. Here, we report the therapeutic efficacy of an 8-mer ASO (3UP8i) in two severe models of SMA. While 3UP8i modestly improved survival and function in the more severe Taiwanese SMA model, it dramatically increased survival, improved neuromuscular junction pathology, and tempered cardiac deficits in a new, less severe model of SMA. Our results expand the repertoire of ASO-based compounds for SMA therapy, and for the first time, demonstrate the in vivo efficacy of a short ASO in the context of a human disease.
Figures


References
-
- Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
-
- Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3:97–110. - PubMed
-
- Dubowitz V. Chaos in the classification of SMA: a possible resolution. Neuromuscul Disord. 1995;5:3–5. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

