Phosphates in the Z-DNA dodecamer are flexible, but their P-SAD signal is sufficient for structure solution
- PMID: 25004957
- PMCID: PMC4089481
- DOI: 10.1107/S1399004714004684
Phosphates in the Z-DNA dodecamer are flexible, but their P-SAD signal is sufficient for structure solution
Abstract
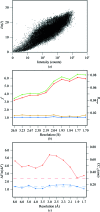
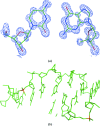
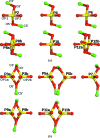
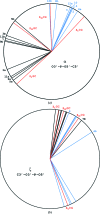
A large number of Z-DNA hexamer duplex structures and a few oligomers of different lengths are available, but here the first crystal structure of the d(CGCGCGCGCGCG)2 dodecameric duplex is presented. Two synchrotron data sets were collected; one was used to solve the structure by the single-wavelength anomalous dispersion (SAD) approach based on the anomalous signal of P atoms, the other set, extending to an ultrahigh resolution of 0.75 Å, served to refine the atomic model to an R factor of 12.2% and an R(free) of 13.4%. The structure consists of parallel duplexes arranged into practically infinitely long helices packed in a hexagonal fashion, analogous to all other known structures of Z-DNA oligomers. However, the dodecamer molecule shows a high level of flexibility, especially of the backbone phosphate groups, with six out of 11 phosphates modeled in double orientations corresponding to the two previously observed Z-DNA conformations: Z(I), with the phosphate groups inclined towards the inside of the helix, and Z(II), with the phosphate groups rotated towards the outside of the helix.
Keywords: Z-DNA dodecamer; Z-DNA structure; flexibility of phosphate groups; phosphorus SAD phasing; ultrahigh resolution.
Figures

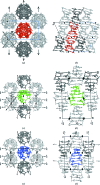

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

