Epigenetic silencing of genes and microRNAs within the imprinted Dlk1-Dio3 region at human chromosome 14.32 in giant cell tumor of bone
- PMID: 25005035
- PMCID: PMC4101709
- DOI: 10.1186/1471-2407-14-495
Epigenetic silencing of genes and microRNAs within the imprinted Dlk1-Dio3 region at human chromosome 14.32 in giant cell tumor of bone
Abstract
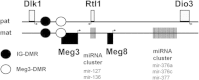
Background: Growing evidence exists that the neoplastic stromal cell population (GCTSC) within giant cell tumors (GCT) originates from mesenchymal stem cells (MSC). In a previous study we identified a microRNA signature that differentiates between these cell types. Five differentially expressed microRNAs are located within the Dlk1-Dio3 region on chromosome 14. Aberrant regulation within this region is known to influence cell growth, differentiation and the development of cancer. The aim of this study was to elucidate the involvement of deregulations within the Dlk1-Dio3 region in GCT pathogenesis.
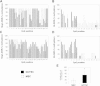
Methods: Quantitative gene and microRNA expression analyses were performed on GCTSCs and MSCs with or without treatment with epigenetic modifiers. Methylation analysis of differentially methylated regions was performed by bisulfite sequencing.
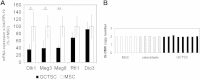
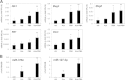
Results: In addition to microRNA silencing we detected a significant downregulation of Dlk1, Meg3 and Meg8 in GCTSCs compared to MSCs. DNA methylation analyses of the Meg3-DMR and IG-DMR revealed a frequent hypermethylation within the IG-DMR in GCTs. Epigenetic modification could restore expression of some but not all analyzed genes and microRNAs suggesting further regulatory mechanisms.
Conclusion: Epigenetic silencing of genes and microRNAs within the Dlk1-Dio3 region is a common event in GCTSCs, in part mediated by hypermethylation within the IG-DMR. The identified genes, micro RNAs and microRNA target genes might be valuable targets for the development of improved strategies for GCT diagnosis and therapy.
Figures


References
-
- Gupta R, Seethalakshmi V, Jambhekar NA, Prabhudesai S, Merchant N, Puri A, Agarwal M. Clinicopathologic profile of 470 giant cell tumors of bone from a cancer hospital in western India. Ann Diagn Pathol. 2008;12:239–248. - PubMed
-
- Wulling M, Engels C, Jesse N, Werner M, Delling G, Kaiser E. The nature of giant cell tumor of bone. J Cancer Res Clin Oncol. 2001;127:467–474. - PubMed
-
- Gamberi G, Benassi MS, Bohling T, Ragazzini P, Molendini L, Sollazzo MR, Merli M, Ferrari C, Magagnoli G, Bertoni F, Picci P. Prognostic relevance of C-myc gene expression in giant-cell tumor of bone. J Orthop Res. 1998;16:1–7. - PubMed
-
- Osaka S, Toriyama M, Taira K, Sano S, Saotome K. Analysis of giant cell tumor of bone with pulmonary metastases. Clin Orthop Relat Res. 1997;335:253–261. - PubMed
-
- Capanna R, Fabbri N, Bettelli G. Curettage of giant cell tumor of bone. The effect of surgical technique and adjuvants on local recurrence rate. Chir Organi Mov. 1990;75:206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

