Recombinant production, crystallization and X-ray crystallographic structure determination of peptidyl-tRNA hydrolase from Salmonella typhimurium
- PMID: 25005080
- PMCID: PMC4089523
- DOI: 10.1107/S2053230X14009893
Recombinant production, crystallization and X-ray crystallographic structure determination of peptidyl-tRNA hydrolase from Salmonella typhimurium
Abstract
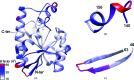
Peptidyl-tRNA hydrolase (Pth; EC 3.1.1.29) from the pathogenic bacterium Salmonella typhimurium has been cloned, expressed in Escherichia coli and crystallized for X-ray analysis. Crystals were grown using hanging-drop vapor diffusion against a reservoir solution consisting of 0.03 M citric acid, 0.05 M bis-tris propane, 1% glycerol, 3% sucrose, 25% PEG 6000 pH 7.6. Crystals were used to obtain the three-dimensional structure of the native protein at 1.6 Å resolution. The structure was determined by molecular replacement of the crystallographic data processed in space group P2₁2₁2₁ with unit-cell parameters a=62.1, b=64.9, c=110.5 Å, α=β=γ=90°. The asymmetric unit of the crystallographic lattice was composed of two copies of the enzyme molecule with a 51% solvent fraction, corresponding to a Matthews coefficient of 2.02 Å3 Da(-1). The structural coordinates reported serve as a foundation for computational and structure-guided efforts towards novel small-molecule Pth1 inhibitors and potential antibacterial development.
Keywords: Salmonella typhimurium; peptidyl-tRNA; peptidyl-tRNA hydrolase 1.
Figures


Similar articles
-
Recombinant production, crystallization and X-ray crystallographic structure determination of the peptidyl-tRNA hydrolase of Pseudomonas aeruginosa.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Dec 1;68(Pt 12):1472-6. doi: 10.1107/S1744309112045770. Epub 2012 Nov 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 23192026 Free PMC article.
-
Crystallization and preliminary X-ray analysis of peptidyl-tRNA hydrolase from Thermus thermophilus HB8.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Mar 1;69(Pt 3):332-5. doi: 10.1107/S1744309113003424. Epub 2013 Feb 27. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23519816 Free PMC article.
-
Structural and functional characterization of peptidyl-tRNA hydrolase from Klebsiella pneumoniae.Biochim Biophys Acta Proteins Proteom. 2021 Jan;1869(1):140554. doi: 10.1016/j.bbapap.2020.140554. Epub 2020 Oct 15. Biochim Biophys Acta Proteins Proteom. 2021. PMID: 33068756
-
Crystallization and preliminary X-ray analysis of peptidyl-tRNA hydrolase from Escherichia coli in complex with the acceptor-TΨC domain of tRNA.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Dec 1;67(Pt 12):1566-9. doi: 10.1107/S1744309111038383. Epub 2011 Nov 26. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 22139168 Free PMC article.
-
Unveiling the Druggable Landscape of Bacterial Peptidyl tRNA Hydrolase: Insights into Structure, Function, and Therapeutic Potential.Biomolecules. 2024 Jun 7;14(6):668. doi: 10.3390/biom14060668. Biomolecules. 2024. PMID: 38927071 Free PMC article. Review.
Cited by
-
Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection.Nat Commun. 2018 May 18;9(1):1993. doi: 10.1038/s41467-018-04472-6. Nat Commun. 2018. PMID: 29777131 Free PMC article.
-
Small Molecule Docking Supports Broad and Narrow Spectrum Potential for the Inhibition of the Novel Antibiotic Target Bacterial Pth1.Antibiotics (Basel). 2016 May 10;5(2):16. doi: 10.3390/antibiotics5020016. Antibiotics (Basel). 2016. PMID: 27171117 Free PMC article.
-
Natural Product Inhibition and Enzyme Kinetics Related to Phylogenetic Characterization for Bacterial Peptidyl-tRNA Hydrolase 1.Molecules. 2021 Apr 15;26(8):2281. doi: 10.3390/molecules26082281. Molecules. 2021. PMID: 33920799 Free PMC article.
-
Binding mode between peptidyl-tRNA hydrolase and the peptidyl-A76 moiety of the substrate.J Biol Chem. 2025 Apr;301(4):108385. doi: 10.1016/j.jbc.2025.108385. Epub 2025 Mar 4. J Biol Chem. 2025. PMID: 40049414 Free PMC article.
-
Unraveling the stereochemical and dynamic aspects of the catalytic site of bacterial peptidyl-tRNA hydrolase.RNA. 2017 Feb;23(2):202-216. doi: 10.1261/rna.057620.116. Epub 2016 Nov 10. RNA. 2017. PMID: 28096445 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

