Stochastic NANOG fluctuations allow mouse embryonic stem cells to explore pluripotency
- PMID: 25005472
- PMCID: PMC6517831
- DOI: 10.1242/dev.108910
Stochastic NANOG fluctuations allow mouse embryonic stem cells to explore pluripotency
Abstract
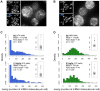
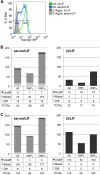
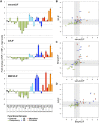
Heterogeneous expression of the transcription factor NANOG has been linked to the existence of various functional states in pluripotent stem cells. This heterogeneity seems to arise from fluctuations of Nanog expression in individual cells, but a thorough characterization of these fluctuations and their impact on the pluripotent state is still lacking. Here, we have used a novel fluorescent reporter to investigate the temporal dynamics of NANOG expression in mouse embryonic stem cells (mESCs), and to dissect the lineage potential of mESCs at different NANOG states. Our results show that stochastic NANOG fluctuations are widespread in mESCs, with essentially all expressing cells showing fluctuations in NANOG levels, even when cultured in ground-state conditions (2i media). We further show that fluctuations have similar kinetics when mESCs are cultured in standard conditions (serum plus leukemia inhibitory factor) or ground-state conditions, implying that NANOG fluctuations are inherent to the pluripotent state. We have then compared the developmental potential of low-NANOG and high-NANOG mESCs, grown in different conditions, and confirm that mESCs are more susceptible to enter differentiation at the low-NANOG state. Further analysis by gene expression profiling reveals that low-NANOG cells have marked expression of lineage-affiliated genes, with variable profiles according to the signalling environment. By contrast, high-NANOG cells show a more stable expression profile in different environments, with minimal expression of lineage markers. Altogether, our data support a model in which stochastic NANOG fluctuations provide opportunities for mESCs to explore multiple lineage options, modulating their probability to change functional state.
Keywords: Gene expression heterogeneity; Lineage priming; Nanog; Pluripotency; Stem cells.
© 2014. Published by The Company of Biologists Ltd.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

