The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin
- PMID: 25006036
- PMCID: PMC4109307
- DOI: 10.7554/eLife.03300
The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin
Abstract
Influenza is notable for its evolutionary capacity to escape immunity targeting the viral hemagglutinin. We used deep mutational scanning to examine the extent to which a high inherent mutational tolerance contributes to this antigenic evolvability. We created mutant viruses that incorporate most of the ≈10(4) amino-acid mutations to hemagglutinin from A/WSN/1933 (H1N1) influenza. After passaging these viruses in tissue culture to select for functional variants, we used deep sequencing to quantify mutation frequencies before and after selection. These data enable us to infer the preference for each amino acid at each site in hemagglutinin. These inferences are consistent with existing knowledge about the protein's structure and function, and can be used to create a model that describes hemagglutinin's evolution far better than existing phylogenetic models. We show that hemagglutinin has a high inherent tolerance for mutations at antigenic sites, suggesting that this is one factor contributing to influenza's antigenic evolution.
Keywords: antigenic evolution; deep mutational scanning; evolutionary biology; evolvability; genomics; hemagglutinin; infectious disease; influenza; microbiology; phylogenetics; viruses.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

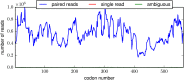
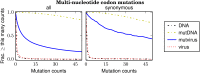





Similar articles
-
Complete mapping of viral escape from neutralizing antibodies.PLoS Pathog. 2017 Mar 13;13(3):e1006271. doi: 10.1371/journal.ppat.1006271. eCollection 2017 Mar. PLoS Pathog. 2017. PMID: 28288189 Free PMC article.
-
Mapping person-to-person variation in viral mutations that escape polyclonal serum targeting influenza hemagglutinin.Elife. 2019 Aug 27;8:e49324. doi: 10.7554/eLife.49324. Elife. 2019. PMID: 31452511 Free PMC article.
-
The Influenza B Virus Hemagglutinin Head Domain Is Less Tolerant to Transposon Mutagenesis than That of the Influenza A Virus.J Virol. 2018 Jul 31;92(16):e00754-18. doi: 10.1128/JVI.00754-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29899093 Free PMC article.
-
Mapping the Evolutionary Potential of RNA Viruses.Cell Host Microbe. 2018 Apr 11;23(4):435-446. doi: 10.1016/j.chom.2018.03.012. Cell Host Microbe. 2018. PMID: 29649440 Free PMC article. Review.
-
Molecular determinants of protein evolvability.Trends Biochem Sci. 2023 Sep;48(9):751-760. doi: 10.1016/j.tibs.2023.05.009. Epub 2023 Jun 15. Trends Biochem Sci. 2023. PMID: 37330341 Review.
Cited by
-
Adaptation of turnip mosaic potyvirus to a specific niche reduces its genetic and environmental robustness.Virus Evol. 2020 May 20;6(2):veaa041. doi: 10.1093/ve/veaa041. eCollection 2020 Jul. Virus Evol. 2020. PMID: 32782826 Free PMC article.
-
Site-Specific Amino Acid Preferences Are Mostly Conserved in Two Closely Related Protein Homologs.Mol Biol Evol. 2015 Nov;32(11):2944-60. doi: 10.1093/molbev/msv167. Epub 2015 Jul 29. Mol Biol Evol. 2015. PMID: 26226986 Free PMC article.
-
The past, present and future of RNA respiratory viruses: influenza and coronaviruses.Pathog Dis. 2020 Oct 7;78(7):ftaa046. doi: 10.1093/femspd/ftaa046. Pathog Dis. 2020. PMID: 32860686 Free PMC article. Review.
-
ProteinGym: Large-Scale Benchmarks for Protein Design and Fitness Prediction.bioRxiv [Preprint]. 2023 Dec 8:2023.12.07.570727. doi: 10.1101/2023.12.07.570727. bioRxiv. 2023. PMID: 38106144 Free PMC article. Preprint.
-
Detection and sequence/structure mapping of biophysical constraints to protein variation in saturated mutational libraries and protein sequence alignments with a dedicated server.BMC Bioinformatics. 2016 Jun 17;17(1):242. doi: 10.1186/s12859-016-1124-4. BMC Bioinformatics. 2016. PMID: 27315797 Free PMC article.
References
-
- Adey A, Morrison HG, Xun X, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, Shendure J. 2010. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biology 11:R119. doi: 10.1186/gb-2010-11-12-r119 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

