The female urinary microbiome: a comparison of women with and without urgency urinary incontinence
- PMID: 25006228
- PMCID: PMC4161260
- DOI: 10.1128/mBio.01283-14
The female urinary microbiome: a comparison of women with and without urgency urinary incontinence
Abstract
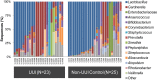
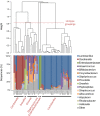
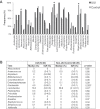
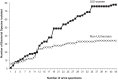
Bacterial DNA and live bacteria have been detected in human urine in the absence of clinical infection, challenging the prevailing dogma that urine is normally sterile. Urgency urinary incontinence (UUI) is a poorly understood urinary condition characterized by symptoms that overlap urinary infection, including urinary urgency and increased frequency with urinary incontinence. The recent discovery of the urinary microbiome warrants investigation into whether bacteria contribute to UUI. In this study, we used 16S rRNA gene sequencing to classify bacterial DNA and expanded quantitative urine culture (EQUC) techniques to isolate live bacteria in urine collected by using a transurethral catheter from women with UUI and, in comparison, a cohort without UUI. For these cohorts, we demonstrated that the UUI and non-UUI urinary microbiomes differ by group based on both sequence and culture evidences. Compared to the non-UUI microbiome, sequencing experiments revealed that the UUI microbiome was composed of increased Gardnerella and decreased Lactobacillus. Nine genera (Actinobaculum, Actinomyces, Aerococcus, Arthrobacter, Corynebacterium, Gardnerella, Oligella, Staphylococcus, and Streptococcus) were more frequently cultured from the UUI cohort. Although Lactobacillus was isolated from both cohorts, distinctions existed at the species level, with Lactobacillus gasseri detected more frequently in the UUI cohort and Lactobacillus crispatus most frequently detected in controls. Combined, these data suggest that potentially important differences exist in the urinary microbiomes of women with and without UUI, which have strong implications in prevention, diagnosis, or treatment of UUI. Importance: New evidence indicates that the human urinary tract contains microbial communities; however, the role of these communities in urinary health remains to be elucidated. Urgency urinary incontinence (UUI) is a highly prevalent yet poorly understood urinary condition characterized by urgency, frequency, and urinary incontinence. Given the significant overlap of UUI symptoms with those of urinary tract infections, it is possible that UUI may have a microbial component. We compared the urinary microbiomes of women affected by UUI to those of a comparison group without UUI, using both high-throughput sequencing and extended culture techniques. We identified statistically significant differences in the frequency and abundance of bacteria present. These differences suggest a potential role for the urinary microbiome in female urinary health.
Trial registration: ClinicalTrials.gov NCT01642277.
Copyright © 2014 Pearce et al.
Figures
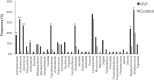
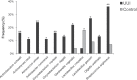
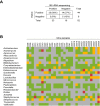
Comment in
-
Incontinence: A role for the microbiome in urge incontinence?Nat Rev Urol. 2014 Sep;11(9):486. doi: 10.1038/nrurol.2014.186. Epub 2014 Jul 29. Nat Rev Urol. 2014. PMID: 25069736 No abstract available.
References
-
- Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ, Huang ST, Ljungberg I, Sprague BM, Lucas SK, Torralba M, Nelson KE, Groah SL. 2012. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J. Transl. Med. 10:174. 10.1186/1479-5876-10-174 - DOI - PMC - PubMed
-
- Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, Brubaker L, Gai X, Wolfe AJ, Schreckenberger PC. 2014. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J. Clin. Microbiol. 52:871–876. 10.1128/JCM.02876-13 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
