Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing
- PMID: 25006232
- PMCID: PMC4161252
- DOI: 10.1128/mBio.01442-14
Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing
Abstract
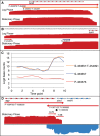
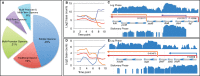
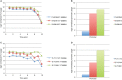
We analyzed the transcriptome of Escherichia coli K-12 by strand-specific RNA sequencing at single-nucleotide resolution during steady-state (logarithmic-phase) growth and upon entry into stationary phase in glucose minimal medium. To generate high-resolution transcriptome maps, we developed an organizational schema which showed that in practice only three features are required to define operon architecture: the promoter, terminator, and deep RNA sequence read coverage. We precisely annotated 2,122 promoters and 1,774 terminators, defining 1,510 operons with an average of 1.98 genes per operon. Our analyses revealed an unprecedented view of E. coli operon architecture. A large proportion (36%) of operons are complex with internal promoters or terminators that generate multiple transcription units. For 43% of operons, we observed differential expression of polycistronic genes, despite being in the same operons, indicating that E. coli operon architecture allows fine-tuning of gene expression. We found that 276 of 370 convergent operons terminate inefficiently, generating complementary 3' transcript ends which overlap on average by 286 nucleotides, and 136 of 388 divergent operons have promoters arranged such that their 5' ends overlap on average by 168 nucleotides. We found 89 antisense transcripts of 397-nucleotide average length, 7 unannotated transcripts within intergenic regions, and 18 sense transcripts that completely overlap operons on the opposite strand. Of 519 overlapping transcripts, 75% correspond to sequences that are highly conserved in E. coli (>50 genomes). Our data extend recent studies showing unexpected transcriptome complexity in several bacteria and suggest that antisense RNA regulation is widespread. Importance: We precisely mapped the 5' and 3' ends of RNA transcripts across the E. coli K-12 genome by using a single-nucleotide analytical approach. Our resulting high-resolution transcriptome maps show that ca. one-third of E. coli operons are complex, with internal promoters and terminators generating multiple transcription units and allowing differential gene expression within these operons. We discovered extensive antisense transcription that results from more than 500 operons, which fully overlap or extensively overlap adjacent divergent or convergent operons. The genomic regions corresponding to these antisense transcripts are highly conserved in E. coli (including Shigella species), although it remains to be proven whether or not they are functional. Our observations of features unearthed by single-nucleotide transcriptome mapping suggest that deeper layers of transcriptional regulation in bacteria are likely to be revealed in the future.
Copyright © 2014 Conway et al.
Figures


References
-
- Lehman IR, Bessman MJ, Simms ES, Kornberg A. 1958. Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli. J. Biol. Chem. 233:163–170 - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
