Identification of multifaceted binding modes for pyrin and ASC pyrin domains gives insights into pyrin inflammasome assembly
- PMID: 25006247
- PMCID: PMC4156052
- DOI: 10.1074/jbc.M114.553305
Identification of multifaceted binding modes for pyrin and ASC pyrin domains gives insights into pyrin inflammasome assembly
Abstract
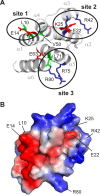
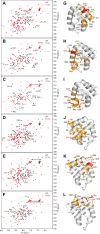
Inflammasomes are macromolecular complexes that mediate inflammatory and cell death responses to pathogens and cellular stress signals. Dysregulated inflammasome activation is associated with autoinflammatory syndromes and several common diseases. During inflammasome assembly, oligomerized cytosolic pattern recognition receptors recruit procaspase-1 and procaspase-8 via the adaptor protein ASC. Inflammasome assembly is mediated by pyrin domains (PYDs) and caspase recruitment domains, which are protein interaction domains of the death fold superfamily. However, the molecular details of their interactions are poorly understood. We have studied the interaction between ASC and pyrin PYDs that mediates ASC recruitment to the pyrin inflammasome, which is implicated in the pathogenesis of familial Mediterranean fever. We demonstrate that both the ASC and pyrin PYDs have multifaceted binding modes, involving three sites on pyrin PYD and two sites on ASC PYD. Molecular docking of pyrin-ASC PYD complexes showed that pyrin PYD can simultaneously interact with up to three ASC PYDs. Furthermore, ASC PYD can self-associate and interact with pyrin, consistent with previous reports that pyrin promotes ASC clustering to form a proinflammatory complex. Finally, the effects of familial Mediterranean fever-associated mutations, R42W and A89T, on structural and functional properties of pyrin PYD were investigated. The R42W mutation had a significant effect on structure and increased stability. Although the R42W mutant exhibited reduced interaction with ASC, it also bound less to the pyrin B-box domain responsible for autoinhibition and hence may be constitutively active. Our data give new insights into the binding modes of PYDs and inflammasome architecture.
Keywords: Death Domain; Familial Mediterranean Fever; Inflammasome; Inflammation; Innate Immunity; Mutagenesis; Pattern Recognition Receptor (PRR); Protein Complex; Protein-Protein Interaction; Pyrin.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






Similar articles
-
Multiple binding sites on the pyrin domain of ASC protein allow self-association and interaction with NLRP3 protein.J Biol Chem. 2012 Dec 7;287(50):41732-43. doi: 10.1074/jbc.M112.381228. Epub 2012 Oct 12. J Biol Chem. 2012. PMID: 23066025 Free PMC article.
-
The Inflammasome Adaptor ASC Induces Procaspase-8 Death Effector Domain Filaments.J Biol Chem. 2015 Dec 4;290(49):29217-30. doi: 10.1074/jbc.M115.687731. Epub 2015 Oct 14. J Biol Chem. 2015. PMID: 26468282 Free PMC article.
-
Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly.J Biol Chem. 2013 May 10;288(19):13225-35. doi: 10.1074/jbc.M113.468033. Epub 2013 Mar 25. J Biol Chem. 2013. PMID: 23530044 Free PMC article.
-
Inhibiting the inflammasome: one domain at a time.Immunol Rev. 2015 May;265(1):205-16. doi: 10.1111/imr.12290. Immunol Rev. 2015. PMID: 25879295 Free PMC article. Review.
-
Structural mechanisms of inflammasome assembly.FEBS J. 2015 Feb;282(3):435-44. doi: 10.1111/febs.13133. Epub 2014 Nov 21. FEBS J. 2015. PMID: 25354325 Free PMC article. Review.
Cited by
-
Update on Pyrin Functions and Mechanisms of Familial Mediterranean Fever.Front Microbiol. 2016 Mar 31;7:456. doi: 10.3389/fmicb.2016.00456. eCollection 2016. Front Microbiol. 2016. PMID: 27066000 Free PMC article. Review.
-
NLRP3 inflammasome: structure, mechanism, drug-induced organ toxicity, therapeutic strategies, and future perspectives.RSC Med Chem. 2025 May 13. doi: 10.1039/d5md00167f. Online ahead of print. RSC Med Chem. 2025. PMID: 40370650 Free PMC article. Review.
-
Transmissible gastroenteritis virus infection induces NF-κB activation through RLR-mediated signaling.Virology. 2017 Jul;507:170-178. doi: 10.1016/j.virol.2017.04.024. Epub 2017 Apr 24. Virology. 2017. PMID: 28448848 Free PMC article.
-
TRIMs: Generalists Regulating the NLRP3 Inflammasome Signaling Pathway.DNA Cell Biol. 2022 Mar;41(3):262-275. doi: 10.1089/dna.2021.0943. Epub 2022 Feb 18. DNA Cell Biol. 2022. PMID: 35180350 Free PMC article. Review.
-
Assembly and regulation of ASC specks.Cell Mol Life Sci. 2017 Apr;74(7):1211-1229. doi: 10.1007/s00018-016-2396-6. Epub 2016 Oct 19. Cell Mol Life Sci. 2017. PMID: 27761594 Free PMC article. Review.
References
-
- Lamkanfi M., Dixit V. M. (2012) Inflammasomes and their roles in health and disease. Annu. Rev. Cell Dev. Biol. 28, 137–161 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

