Function and Evolution of the Sox Multienzyme Complex in the Marine Gammaproteobacterium Congregibacter litoralis
- PMID: 25006520
- PMCID: PMC4003848
- DOI: 10.1155/2014/597418
Function and Evolution of the Sox Multienzyme Complex in the Marine Gammaproteobacterium Congregibacter litoralis
Abstract
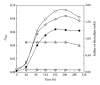
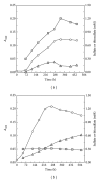
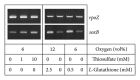
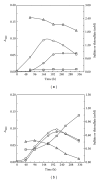
Core sets of sox genes were detected in several genome sequenced members of the environmental important OM60/NOR5 clade of marine gammaproteobacteria. However, emendation of media with thiosulfate did not result in stimulation of growth in two of these strains and cultures of Congregibacter litoralis DSM 17192(T) did not oxidize thiosulfate to sulfate in concentrations of one mmol L(-1) or above. On the other hand, a significant production of sulfate was detected upon growth with the organic sulfur compounds, cysteine and glutathione. It was found that degradation of glutathione resulted in the formation of submillimolar amounts of thiosulfate in the closely related sox-negative strain Chromatocurvus halotolerans DSM 23344(T). It is proposed that the Sox multienzyme complex in Congregibacter litoralis and related members of the OM60/NOR5 clade is adapted to the oxidation of submillimolar amounts of thiosulfate and nonfunctional at higher concentrations of reduced inorganic sulfur compounds. Pelagic bacteria thriving in the oxic zones of marine environments may rarely encounter amounts of thiosulfate, which would allow its utilization as electron donor for lithoautotrophic or mixotrophic growth. Consequently, in evolution the Sox multienzyme complex in some of these bacteria may have been optimized for the effective utilization of trace amounts of thiosulfate generated from the degradation of organic sulfur compounds.
Figures
Similar articles
-
Taxonomy and evolution of bacteriochlorophyll a-containing members of the OM60/NOR5 clade of marine gammaproteobacteria: description of Luminiphilus syltensis gen. nov., sp. nov., reclassification of Haliea rubra as Pseudohaliea rubra gen. nov., comb. nov., and emendation of Chromatocurvus halotolerans.BMC Microbiol. 2013 May 24;13:118. doi: 10.1186/1471-2180-13-118. BMC Microbiol. 2013. PMID: 23705883 Free PMC article.
-
Mixotrophic growth of bacteriochlorophyll a-containing members of the OM60/NOR5 clade of marine gammaproteobacteria is carbon-starvation independent and correlates with the type of carbon source and oxygen availability.BMC Microbiol. 2013 May 24;13:117. doi: 10.1186/1471-2180-13-117. BMC Microbiol. 2013. PMID: 23705861 Free PMC article.
-
Biogeography and phylogeny of the NOR5/OM60 clade of Gammaproteobacteria.Syst Appl Microbiol. 2009 Apr;32(2):124-39. doi: 10.1016/j.syapm.2008.12.001. Syst Appl Microbiol. 2009. PMID: 19216045
-
[Oxidation of inorganic sulfur compounds by obligatory organotrophic bacteria].Mikrobiologiia. 2003 Nov-Dec;72(6):725-39. Mikrobiologiia. 2003. PMID: 14768537 Review. Russian.
-
Inorganic sulfur oxidizing system in green sulfur bacteria.Photosynth Res. 2010 Jun;104(2-3):163-76. doi: 10.1007/s11120-010-9531-2. Epub 2010 Feb 9. Photosynth Res. 2010. PMID: 20143161 Review.
Cited by
-
A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data.Front Microbiol. 2015 Apr 9;6:281. doi: 10.3389/fmicb.2015.00281. eCollection 2015. Front Microbiol. 2015. PMID: 25914684 Free PMC article.
-
The complete genome sequence of Rhodobaca barguzinensis alga05 (DSM 19920) documents its adaptation for life in soda lakes.Extremophiles. 2018 Nov;22(6):839-849. doi: 10.1007/s00792-018-1041-8. Epub 2018 Jul 18. Extremophiles. 2018. PMID: 30022245
-
Phylogenomic Analyses of Members of the Widespread Marine Heterotrophic Genus Pseudovibrio Suggest Distinct Evolutionary Trajectories and a Novel Genus, Polycladidibacter gen. nov.Appl Environ Microbiol. 2020 Feb 3;86(4):e02395-19. doi: 10.1128/AEM.02395-19. Print 2020 Feb 3. Appl Environ Microbiol. 2020. PMID: 31811036 Free PMC article.
-
Uncovering multi-faceted taxonomic and functional diversity of soil bacteriomes in tropical Southeast Asian countries.Sci Rep. 2021 Jan 12;11(1):582. doi: 10.1038/s41598-020-79786-x. Sci Rep. 2021. PMID: 33436774 Free PMC article.
-
Metabolic Versatility of the Family Halieaceae Revealed by the Genomics of Novel Cultured Isolates.Microbiol Spectr. 2023 Mar 14;11(2):e0387922. doi: 10.1128/spectrum.03879-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36916946 Free PMC article.
References
-
- Yan S, Fuchs BM, Lenk S, et al. Biogeography and phylogeny of the NOR5/OM60 clade of Gammaproteobacteria . Systematic and Applied Microbiology. 2009;32(2):124–139. - PubMed
-
- Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J. Prokaryotic sulfur oxidation. Current Opinion in Microbiology. 2005;8(3):253–259. - PubMed
-
- Meyer B, Imhoff JF, Kuever J. Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria—evolution of the Sox sulfur oxidation enzyme system. Environmental Microbiology. 2007;9(12):2957–2977. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

