Atg6/UVRAG/Vps34-containing lipid kinase complex is required for receptor downregulation through endolysosomal degradation and epithelial polarity during Drosophila wing development
- PMID: 25006588
- PMCID: PMC4074780
- DOI: 10.1155/2014/851349
Atg6/UVRAG/Vps34-containing lipid kinase complex is required for receptor downregulation through endolysosomal degradation and epithelial polarity during Drosophila wing development
Abstract
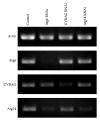
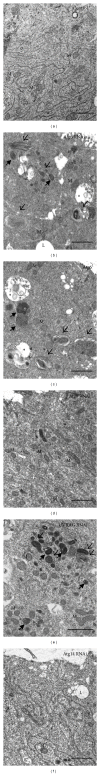
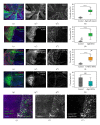
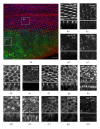
Atg6 (Beclin 1 in mammals) is a core component of the Vps34 PI3K (III) complex, which promotes multiple vesicle trafficking pathways. Atg6 and Vps34 form two distinct PI3K (III) complexes in yeast and mammalian cells, either with Atg14 or with UVRAG. The functions of these two complexes are not entirely clear, as both Atg14 and UVRAG have been suggested to regulate both endocytosis and autophagy. In this study, we performed a microscopic analysis of UVRAG, Atg14, or Atg6 loss-of-function cells in the developing Drosophila wing. Both autophagy and endocytosis are seriously impaired and defective endolysosomes accumulate upon loss of Atg6. We show that Atg6 is required for the downregulation of Notch and Wingless signaling pathways; thus it is essential for normal wing development. Moreover, the loss of Atg6 impairs cell polarity. Atg14 depletion results in autophagy defects with no effect on endocytosis or cell polarity, while the silencing of UVRAG phenocopies all but the autophagy defect of Atg6 depleted cells. Thus, our results indicate that the UVRAG-containing PI3K (III) complex is required for receptor downregulation through endolysosomal degradation and for the establishment of proper cell polarity in the developing wing, while the Atg14-containing complex is involved in autophagosome formation.
Figures







Similar articles
-
Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG.Mol Biol Cell. 2008 Dec;19(12):5360-72. doi: 10.1091/mbc.e08-01-0080. Epub 2008 Oct 8. Mol Biol Cell. 2008. PMID: 18843052 Free PMC article.
-
VPS38/UVRAG and ATG14, the variant regulatory subunits of the ATG6/Beclin1-PI3K complexes, are crucial for the biogenesis of the yolk organelles and are transcriptionally regulated in the oocytes of the vector Rhodnius prolixus.PLoS Negl Trop Dis. 2021 Sep 7;15(9):e0009760. doi: 10.1371/journal.pntd.0009760. eCollection 2021 Sep. PLoS Negl Trop Dis. 2021. PMID: 34492013 Free PMC article.
-
Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes.Autophagy. 2009 May;5(4):534-6. doi: 10.4161/auto.5.4.8062. Epub 2009 May 2. Autophagy. 2009. PMID: 19223761
-
Forces shaping the Drosophila wing.Mech Dev. 2017 Apr;144(Pt A):23-32. doi: 10.1016/j.mod.2016.10.003. Epub 2016 Oct 23. Mech Dev. 2017. PMID: 27784612 Review.
-
Activation Mechanisms of the VPS34 Complexes.Cells. 2021 Nov 11;10(11):3124. doi: 10.3390/cells10113124. Cells. 2021. PMID: 34831348 Free PMC article. Review.
Cited by
-
BECLIN1: Protein Structure, Function and Regulation.Cells. 2021 Jun 17;10(6):1522. doi: 10.3390/cells10061522. Cells. 2021. PMID: 34204202 Free PMC article. Review.
-
Condition-dependent functional shift of two Drosophila Mtmr lipid phosphatases in autophagy control.Autophagy. 2021 Dec;17(12):4010-4028. doi: 10.1080/15548627.2021.1899681. Epub 2021 Mar 28. Autophagy. 2021. PMID: 33779490 Free PMC article.
-
Autophagic UVRAG Promotes UV-Induced Photolesion Repair by Activation of the CRL4(DDB2) E3 Ligase.Mol Cell. 2016 May 19;62(4):507-19. doi: 10.1016/j.molcel.2016.04.014. Mol Cell. 2016. PMID: 27203177 Free PMC article.
-
Beclin 1 Promotes Endosome Recruitment of Hepatocyte Growth Factor Tyrosine Kinase Substrate to Suppress Tumor Proliferation.Cancer Res. 2020 Jan 15;80(2):249-262. doi: 10.1158/0008-5472.CAN-19-1555. Epub 2019 Nov 19. Cancer Res. 2020. PMID: 31744816 Free PMC article.
-
Neuronal-specific septin-3 binds Atg8/LC3B, accumulates and localizes to autophagosomes during induced autophagy.Cell Mol Life Sci. 2022 Aug 6;79(9):471. doi: 10.1007/s00018-022-04488-8. Cell Mol Life Sci. 2022. PMID: 35932293 Free PMC article.
References
-
- Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annual Review of Cell and Developmental Biology. 2011;27:107–132. - PubMed
-
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. - PubMed
-
- Hegedűs K, Takáts S, Kovács AL, Juhász G. Evolutionarily conserved role and physiological relevance of a STX17/Syx17 (syntaxin 17)-containing SNARE complex in autophagosome fusion with endosomes and lysosomes. Autophagy. 2013;9:1642–1646. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

