Non-circadian expression masking clock-driven weak transcription rhythms in U2OS cells
- PMID: 25007071
- PMCID: PMC4090172
- DOI: 10.1371/journal.pone.0102238
Non-circadian expression masking clock-driven weak transcription rhythms in U2OS cells
Abstract
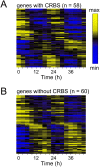
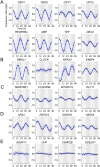
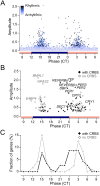
U2OS cells harbor a circadian clock but express only a few rhythmic genes in constant conditions. We identified 3040 binding sites of the circadian regulators BMAL1, CLOCK and CRY1 in the U2OS genome. Most binding sites even in promoters do not correlate with detectable rhythmic transcript levels. Luciferase fusions reveal that the circadian clock supports robust but low amplitude transcription rhythms of representative promoters. However, rhythmic transcription of these potentially clock-controlled genes is masked by non-circadian transcription that overwrites the weaker contribution of the clock in constant conditions. Our data suggest that U2OS cells harbor an intrinsically rather weak circadian oscillator. The oscillator has the potential to regulate a large number of genes. The contribution of circadian versus non-circadian transcription is dependent on the metabolic state of the cell and may determine the apparent complexity of the circadian transcriptome.
Conflict of interest statement
Figures


References
-
- Hogenesch JB, Gu Y-Z, Jain S, Bradfield CA (1998) The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proceedings of the National Academy of Sciences of the United States of America 95: 5474–5479 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=20401&tool=pmc.... - PMC - PubMed
-
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, et al.. (1998) Role of the CLOCK Protein in the Mammalian Circadian Mechanism. Science 280: 1564–1569. Available: http://www.sciencemag.org/cgi/doi/10.1126/science.280.5369.1564. Accessed 2012 March 9. - DOI - PubMed
-
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, et al. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205 Available: http://www.ncbi.nlm.nih.gov/pubmed/10428031. - PubMed
-
- Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, et al. (1998) Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron 21: 1101–1113 Available: http://www.ncbi.nlm.nih.gov/pubmed/9856465. - PubMed
-
- Preitner N, Damiola F, Zakany J, Duboule D, Albrecht U, et al. (2002) The Orphan Nuclear Receptor REV-ERB a Controls Circadian Transcription within the Positive Limb of the Mammalian Circadian Oscillator. Cell 110: 251–260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

