Surveillance of artemether-lumefantrine associated Plasmodium falciparum multidrug resistance protein-1 gene polymorphisms in Tanzania
- PMID: 25007802
- PMCID: PMC4099215
- DOI: 10.1186/1475-2875-13-264
Surveillance of artemether-lumefantrine associated Plasmodium falciparum multidrug resistance protein-1 gene polymorphisms in Tanzania
Abstract
Background: Resistance to anti-malarials is a major public health problem worldwide. After deployment of artemisinin-based combination therapy (ACT) there have been reports of reduced sensitivity to ACT by malaria parasites in South-East Asia. In Tanzania, artemether-lumefantrine (ALu) is the recommended first-line drug in treatment of uncomplicated malaria. This study surveyed the distribution of the Plasmodium falciparum multidrug resistance protein-1 single nucleotide polymorphisms (SNPs) associated with increased parasite tolerance to ALu, in Tanzania.
Methods: A total of 687 Plasmodium falciparum positive dried blood spots on filter paper and rapid diagnostic test strips collected by finger pricks from patients attending health facilities in six regions of Tanzania mainland between June 2010 and August 2011 were used. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique was used to detect Pfmdr1 SNPs N86Y, Y184F and D1246Y.
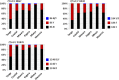
Results: There were variations in the distribution of Pfmdr1 polymorphisms among regions. Tanga region had exceptionally high prevalence of mutant alleles, while Mbeya had the highest prevalence of wild type alleles. The haplotype YFY was exclusively most prevalent in Tanga (29.6%) whereas the NYD haplotype was the most prevalent in all other regions. Excluding Tanga and Mbeya, four, most common Pfmdr1 haplotypes did not vary between the remaining four regions (χ² = 2.3, p = 0.512). The NFD haplotype was the second most prevalent haplotype in all regions, ranging from 17% - 26%.
Conclusion: This is the first country-wide survey on Pfmdr1 mutations associated with ACT resistance. Distribution of individual Pfmdr1 mutations at codons 86, 184 and 1246 varies throughout Tanzanian regions. There is a general homogeneity in distribution of common Pfmdr1 haplotypes reflecting strict implementation of ALu policy in Tanzania with overall prevalence of NFD haplotype ranging from 17 to 26% among other haplotypes. With continuation of ALu as first-line drug this haplotype is expected to keep rising, thus there is need for continued pharmacovigilance studies to monitor any delayed parasite clearance by the drug.
Figures

References
-
- Duraisingh MT, Drakeley CJ, Muller O, Bailey R, Snounou G, Targett GA, Greenwood BM, Warhurst DC. Evidence for selection for the tyrosine-86 allele of the pfmdr 1 gene of Plasmodium falciparum by chloroquine and amodiaquine. Parasitology. 1997;114:205–211. - PubMed
-
- Tinto H, Guekoun L, Zongo I, Guiguemde RT, D’Alessandro U, Ouedraogo JB. Chloroquine-resistance molecular markers (Pfcrt T76 and Pfmdr-1 Y86) and amodiaquine resistance in Burkina Faso. Trop Med Int Health. 2008;13:238–240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

