NMDA receptor structures reveal subunit arrangement and pore architecture
- PMID: 25008524
- PMCID: PMC4263351
- DOI: 10.1038/nature13548
NMDA receptor structures reveal subunit arrangement and pore architecture
Abstract
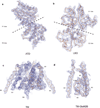
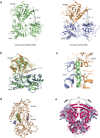
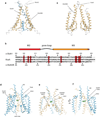
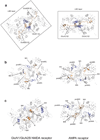
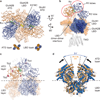
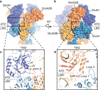
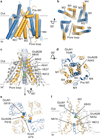
N-methyl-d-aspartate (NMDA) receptors are Hebbian-like coincidence detectors, requiring binding of glycine and glutamate in combination with the relief of voltage-dependent magnesium block to open an ion conductive pore across the membrane bilayer. Despite the importance of the NMDA receptor in the development and function of the brain, a molecular structure of an intact receptor has remained elusive. Here we present X-ray crystal structures of the Xenopus laevis GluN1-GluN2B NMDA receptor with the allosteric inhibitor, Ro25-6981, partial agonists and the ion channel blocker, MK-801. Receptor subunits are arranged in a 1-2-1-2 fashion, demonstrating extensive interactions between the amino-terminal and ligand-binding domains. The transmembrane domains harbour a closed-blocked ion channel, a pyramidal central vestibule lined by residues implicated in binding ion channel blockers and magnesium, and a ∼twofold symmetric arrangement of ion channel pore loops. These structures provide new insights into the architecture, allosteric coupling and ion channel function of NMDA receptors.
Figures




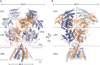

References
-
- Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 2013;14:383–400. - PubMed
-
- Bliss TVP, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. - PubMed
-
- Peery HE, et al. Anti-NMDA receptor encephalitis. The disorder, the diagnosis and the immunobiology. Autoimmun. Rev. 2012;11:863–872. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

