Sequence of events in measles virus replication: role of phosphoprotein-nucleocapsid interactions
- PMID: 25008930
- PMCID: PMC4178853
- DOI: 10.1128/JVI.00664-14
Sequence of events in measles virus replication: role of phosphoprotein-nucleocapsid interactions
Abstract
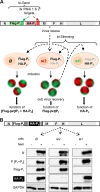
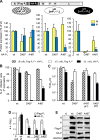
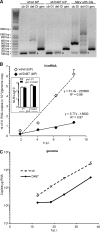
The genome of nonsegmented negative-strand RNA viruses is tightly embedded within a nucleocapsid made of a nucleoprotein (N) homopolymer. To ensure processive RNA synthesis, the viral polymerase L in complex with its cofactor phosphoprotein (P) binds the nucleocapsid that constitutes the functional template. Measles virus P and N interact through two binding sites. While binding of the P amino terminus with the core of N (NCORE) prevents illegitimate encapsidation of cellular RNA, the interaction between their C-terminal domains, P(XD) and N(TAIL) is required for viral RNA synthesis. To investigate the binding dynamics between the two latter domains, the P(XD) F497 residue that makes multiple hydrophobic intramolecular interactions was mutated. Using a quantitative mammalian protein complementation assay and recombinant viruses, we found that an increase in P(XD)-to-N(TAIL) binding strength is associated with a slower transcript accumulation rate and that abolishing the interaction renders the polymerase nonfunctional. The use of a newly developed system allowing conditional expression of wild-type or mutated P genes, revealed that the loss of the P(XD)-N(TAIL) interaction results in reduced transcription by preformed transcriptases, suggesting reduced engagement on the genomic template. These intracellular data indicate that the viral polymerase entry into and progression along its genomic template relies on a protein-protein interaction that serves as a tightly controlled dynamic anchor.
Importance: Mononegavirales have a unique machinery to replicate RNA. Processivity of their polymerase is only achieved when the genome template is entirely embedded into a helical homopolymer of nucleoproteins that constitutes the nucleocapsid. The polymerase binds to the nucleocapsid template through the phosphoprotein. How the polymerase complex enters and travels along the nucleocapsid template to ensure uninterrupted synthesis of up to ∼ 6,700-nucleotide messenger RNAs from six to ten consecutive genes is unknown. Using a quantitative protein complementation assay and a biGene-biSilencing system allowing conditional expression of two P genes copies, the role of the P-to-N interaction in polymerase function was further characterized. We report here a dynamic protein anchoring mechanism that differs from all other known polymerases that rely only onto a sustained and direct binding to their nucleic acid template.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Modulation of Re-initiation of Measles Virus Transcription at Intergenic Regions by PXD to NTAIL Binding Strength.PLoS Pathog. 2016 Dec 9;12(12):e1006058. doi: 10.1371/journal.ppat.1006058. eCollection 2016 Dec. PLoS Pathog. 2016. PMID: 27936158 Free PMC article.
-
Nucleocapsid structure and function.Curr Top Microbiol Immunol. 2009;329:103-28. doi: 10.1007/978-3-540-70523-9_6. Curr Top Microbiol Immunol. 2009. PMID: 19198564 Review.
-
Plasticity in structural and functional interactions between the phosphoprotein and nucleoprotein of measles virus.J Biol Chem. 2012 Apr 6;287(15):11951-67. doi: 10.1074/jbc.M111.333088. Epub 2012 Feb 8. J Biol Chem. 2012. PMID: 22318731 Free PMC article.
-
Crystal Structure of the Measles Virus Nucleoprotein Core in Complex with an N-Terminal Region of Phosphoprotein.J Virol. 2015 Dec 30;90(6):2849-57. doi: 10.1128/JVI.02865-15. J Virol. 2015. PMID: 26719278 Free PMC article.
-
Structural disorder within the replicative complex of measles virus: functional implications.Virology. 2006 Jan 5;344(1):94-110. doi: 10.1016/j.virol.2005.09.025. Virology. 2006. PMID: 16364741 Review.
Cited by
-
Cryo-EM structures of Nipah virus polymerase complex reveal highly varied interactions between L and P proteins among paramyxoviruses.Protein Cell. 2025 Aug 7;16(8):705-723. doi: 10.1093/procel/pwaf014. Protein Cell. 2025. PMID: 39964914 Free PMC article.
-
The Nucleoprotein and Phosphoprotein of Measles Virus.Front Microbiol. 2019 Aug 21;10:1832. doi: 10.3389/fmicb.2019.01832. eCollection 2019. Front Microbiol. 2019. PMID: 31496998 Free PMC article. Review.
-
Breaking the barriers in cancer care: The next generation of herpes simplex virus-based oncolytic immunotherapies for cancer treatment.Mol Ther Oncolytics. 2023 Sep 19;31:100729. doi: 10.1016/j.omto.2023.100729. eCollection 2023 Dec 19. Mol Ther Oncolytics. 2023. PMID: 37841530 Free PMC article. Review.
-
Regulation of NF-κB by the p105-ABIN2-TPL2 complex and RelAp43 during rabies virus infection.PLoS Pathog. 2017 Oct 30;13(10):e1006697. doi: 10.1371/journal.ppat.1006697. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 29084252 Free PMC article.
-
Reduced Nucleoprotein Availability Impairs Negative-Sense RNA Virus Replication and Promotes Host Recognition.J Virol. 2021 Apr 12;95(9):e02274-20. doi: 10.1128/JVI.02274-20. Print 2021 Apr 12. J Virol. 2021. PMID: 33568513 Free PMC article.
References
-
- Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eleouet JF, Rey FA. 2009. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326:1279–1283. 10.1126/science.1177634 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

