Clinicopathologic characteristics and gene expression analyses of non-KRAS 12/13, RAS-mutated metastatic colorectal cancer
- PMID: 25009008
- PMCID: PMC4176451
- DOI: 10.1093/annonc/mdu252
Clinicopathologic characteristics and gene expression analyses of non-KRAS 12/13, RAS-mutated metastatic colorectal cancer
Abstract
Background: KRAS mutations in codons 12 and 13 are present in ∼40% of all colorectal cancers (CRC). Activating mutations in codons 61 and 146 of KRAS and in codons 12, 13, and 61 of NRAS also occur but are less frequent. The clinicopathologic features and gene expression profiles of this latter subpopulation of RAS-mutant colorectal tumors have not yet been clearly defined but in general are treated similarly to those with KRAS 12 or 13 mutations.
Patients and methods: Records of patients with metastatic CRC (mCRC) treated at MD Anderson Cancer Center between December 2000 and August 2012 were reviewed for RAS (KRAS or NRAS) and BRAF mutation status, clinical characteristics, and survival outcomes. To study further with an independent cohort, data from The Cancer Genome Atlas were analyzed to define a gene expression signature for patients whose tumors feature these atypical RAS mutations and explore differences with KRAS 12/13-mutated colorectal tumors.
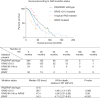
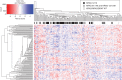
Results: Among the 484 patients reviewed, KRAS 12/13, KRAS 61/146, NRAS, and BRAF mutations were detected in 47.7%, 3.0%, 4.1%, and 7.4%, respectively, of patients who were tested for each of these aberrations. Lung metastases were more common in both the KRAS 12/13-mutated and atypical RAS-mutated cohorts relative to patients with RAS/BRAF wild-type tumors. Gene expression analyses revealed similar patterns regardless of the site of RAS mutation, and in silico functional algorithms predicted that KRAS and NRAS mutations in codons 12, 13, 61, and 146 alter the protein function and drive tumorgenesis.
Conclusions: Clinicopathologic characteristics, survival outcomes, functional impact, and gene expression profiling were similar between patients with KRAS 12/13 and those with NRAS or KRAS 61/146-mutated mCRC. These clinical and bioinformatic findings support the notion that colorectal tumors driven by these RAS mutations are phenotypically similar.
Keywords: KRAS; colorectal cancer; gene expression; next-generation sequencing; survival.
© The Author 2014. Published by Oxford University Press on behalf of the European Society for Medical Oncology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

Similar articles
-
Site-Specific Mutations on KRAS, NRAS, and BRAF Corelate With the Frequency of ctDNA in Colorectal Cancer.Cancer Rep (Hoboken). 2025 Jul;8(7):e70292. doi: 10.1002/cnr2.70292. Cancer Rep (Hoboken). 2025. PMID: 40698441 Free PMC article.
-
EGFR gene gain and PTEN protein expression are favorable prognostic factors in patients with KRAS wild-type metastatic colorectal cancer treated with cetuximab.J Cancer Res Clin Oncol. 2014 May;140(5):737-48. doi: 10.1007/s00432-014-1626-2. Epub 2014 Mar 5. J Cancer Res Clin Oncol. 2014. PMID: 24595598 Free PMC article.
-
Promising biomarkers for predicting the outcomes of patients with KRAS wild-type metastatic colorectal cancer treated with anti-epidermal growth factor receptor monoclonal antibodies: a systematic review with meta-analysis.Int J Cancer. 2013 Oct 15;133(8):1914-25. doi: 10.1002/ijc.28153. Epub 2013 Jul 13. Int J Cancer. 2013. PMID: 23494461
-
The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis.Acta Oncol. 2014 Jul;53(7):852-64. doi: 10.3109/0284186X.2014.895036. Epub 2014 Mar 25. Acta Oncol. 2014. PMID: 24666267
-
KRAS p.G13D mutations are associated with sensitivity to anti-EGFR antibody treatment in colorectal cancer cell lines.J Cancer Res Clin Oncol. 2013 Feb;139(2):201-9. doi: 10.1007/s00432-012-1319-7. Epub 2012 Sep 27. J Cancer Res Clin Oncol. 2013. PMID: 23015072 Free PMC article.
Cited by
-
RGD4C peptide mediates anti-p21Ras scFv entry into tumor cells and produces an inhibitory effect on the human colon cancer cell line SW480.BMC Cancer. 2021 Mar 25;21(1):321. doi: 10.1186/s12885-021-08056-4. BMC Cancer. 2021. PMID: 33765976 Free PMC article.
-
Patterns of Somatic Variants in Colorectal Adenoma and Carcinoma Tissue and Matched Plasma Samples from the Hungarian Oncogenome Program.Cancers (Basel). 2023 Jan 31;15(3):907. doi: 10.3390/cancers15030907. Cancers (Basel). 2023. PMID: 36765865 Free PMC article.
-
Molecular spectrum of KRAS, NRAS, BRAF, PIK3CA, TP53, and APC somatic gene mutations in Arab patients with colorectal cancer: determination of frequency and distribution pattern.J Gastrointest Oncol. 2016 Dec;7(6):882-902. doi: 10.21037/jgo.2016.11.02. J Gastrointest Oncol. 2016. PMID: 28078112 Free PMC article.
-
Colorectal Cancer: From Genetic Landscape to Targeted Therapy.J Oncol. 2021 Jul 6;2021:9918116. doi: 10.1155/2021/9918116. eCollection 2021. J Oncol. 2021. PMID: 34326875 Free PMC article. Review.
-
Prevalence of RAS and BRAF mutations in metastatic colorectal cancer patients by tumor sidedness: A systematic review and meta-analysis.Cancer Med. 2020 Feb;9(3):1044-1057. doi: 10.1002/cam4.2747. Epub 2019 Dec 19. Cancer Med. 2020. PMID: 31856410 Free PMC article.
References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. - PubMed
-
- Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the multicenter ‘RASCAL’ study. J Natl Cancer Inst. 1998;90:675–684. - PubMed
-
- Scheffzek K, Ahmadian MR, Kabsch W, et al. The Ras–RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. - PubMed
-
- Trahey M, McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238:542–545. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

