Allosteric regulation of rhomboid intramembrane proteolysis
- PMID: 25009246
- PMCID: PMC4195783
- DOI: 10.15252/embj.201488149
Allosteric regulation of rhomboid intramembrane proteolysis
Abstract
Proteolysis within the lipid bilayer is poorly understood, in particular the regulation of substrate cleavage. Rhomboids are a family of ubiquitous intramembrane serine proteases that harbour a buried active site and are known to cleave transmembrane substrates with broad specificity. In vitro gel and Förster resonance energy transfer (FRET)-based kinetic assays were developed to analyse cleavage of the transmembrane substrate psTatA (TatA from Providencia stuartii). We demonstrate significant differences in catalytic efficiency (kcat/K0.5) values for transmembrane substrate psTatA (TatA from Providencia stuartii) cleavage for three rhomboids: AarA from P. stuartii, ecGlpG from Escherichia coli and hiGlpG from Haemophilus influenzae demonstrating that rhomboids specifically recognize this substrate. Furthermore, binding of psTatA occurs with positive cooperativity. Competitive binding studies reveal an exosite-mediated mode of substrate binding, indicating allostery plays a role in substrate catalysis. We reveal that exosite formation is dependent on the oligomeric state of rhomboids, and when dimers are dissociated, allosteric substrate activation is not observed. We present a novel mechanism for specific substrate cleavage involving several dynamic processes including positive cooperativity and homotropic allostery for this interesting class of intramembrane proteases.
Keywords: GlpG; allostery; intramembrane protease; kinetics; rhomboid protease.
© 2014 The Authors.
Figures

Topology of three studied rhomboids. hiGlpG and ecGlpG are the simplest and commonly found in prokaryotes. AarA has 7 TMD similar to rhomboids typically found in eukaryotes.
Hill plots of formation of rhomboid-cleaved psTatA versus psTatA concentration (n = 2, mean ± SD). The product concentration was calculated as the percentage of cleaved substrate from the blot image.
Western blot analysis of rhomboid-mediated cleavage of psTatA. psTatA at the concentration range of 0.5–15 μM was incubated with 0.33 μM (hiGlpG and ecGlpG) or 0.13 μM (AarA) at 37°C for 4 h or 15 min, and the cleavage product (C) was separated from the uncleaved substrate (U) with SDS-Tricine gel and developed with Western blot using anti-His antibodies. The star represents a contaminant band from the substrate sample.
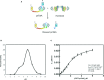
Schematic illustration of the FRET-based cleavage assay. TatA structure determined by NMR (Rodriguez et al, 2013).
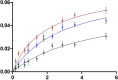
AarA activity dependence on pH. The proteolysis reaction was performed at 37°C for 2 h in the presence of 0.18 μM of rhomboid protease and 2 μM of full-length psTatA-FRET in 150 mM NaCl, 20% glycerol, 0.1% DDM and 2 mM DTT reaction buffer, at pH interval spanning from 2 to 9 with 50 mM of a broad-range pH triple buffer (boric acid:citric acid:phosphate) (Carmody, 1961). The highest initial velocity was calculated to determine the percent activity along the pH gradient. The same pH activity dependence is observed when using a single buffer (Supplementary Fig S3A).
Hill plot of AarA-mediated cleavage of psTatA-FRET. psTatA-FRET at the concentration range from 0.13 to 7 μM was incubated with 0.135 μM of AarA at 37°C for 2 h. Velocities were calculated as described in Materials and Methods (n = 5, mean ± SD). See Table2.

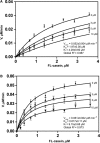
Competitive inhibition of ecGlpG by psTatA, with FL-casein as substrate. The protease reaction between ecGlpG (0.179 μM) with FL-casein (0.179–3.85 μM) was performed in the presence of different concentrations of psTatA (0, 3, 5 and 10 μM). The reaction was started with the protease. Fluorescence emission at 513 nm was measured at 37°C every 5 min for 2 h in a fluorescence microplate reader with an excitation wavelength of 503 nm. Fluorescence detection of each substrate concentration in the presence of corresponding psTatA concentration without enzyme was used as a negative control (n = 2, mean ± SD). Initial velocities were determined for each substrate concentration. Michaelis–Menten plots were subjected to global fit to distinguish the kinetic model and determine the kinetic parameters.
Non-competitive inhibition of AarA by psTatA, with FL-casein as substrate. The assay was conducted, and the kinetic model was determined similar to that for ecGlpG. The concentration of psTatA used is shown beside the corresponding curve.

Comment in
-
Sharpening rhomboid specificity by dimerisation and allostery.EMBO J. 2014 Sep 1;33(17):1847-8. doi: 10.15252/embj.201489373. Epub 2014 Jul 15. EMBO J. 2014. PMID: 25027763 Free PMC article.
References
-
- Ahmedli NB, Gribanova Y, Njoku CC, Naidu A, Young A, Mendoza E, Yamashita CK, Ozgul RK, Johnson JE, Fox DA, Farber DB. Dynamics of the rhomboid-like protein RHBDD2 expression in mouse retina and involvement of its human ortholog in retinitis pigmentosa. J Biol Chem. 2013;288:9742–9754. - PMC - PubMed
-
- Akiyama Y, Maegawa S. Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol Microbiol. 2007;64:1028–1037. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

