SNF5/INI1 deficiency redefines chromatin remodeling complex composition during tumor development
- PMID: 25009291
- PMCID: PMC4233162
- DOI: 10.1158/1541-7786.MCR-14-0005
SNF5/INI1 deficiency redefines chromatin remodeling complex composition during tumor development
Abstract
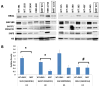
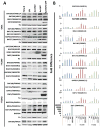
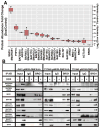
Malignant rhabdoid tumors (MRT), a pediatric cancer that most frequently appears in the kidney and brain, generally lack SNF5 (SMARCB1/INI1), a subunit of the SWI/SNF chromatin-remodeling complex. Recent studies have established that multiple SWI/SNF complexes exist due to the presence or absence of different complex members. Therefore, the effect of SNF5 loss upon SWI/SNF complex formation was investigated in human MRT cells. MRT cells and primary human tumors exhibited reduced levels of many complex proteins. Furthermore, reexpression of SNF5 increased SWI/SNF complex protein levels without concomitant increases in mRNA. Proteomic analysis, using mass spectrometry, of MRT cells before and after SNF5 reexpression indicated the recruitment of different components into the complex along with the expulsion of others. IP-Western blotting confirmed these results and demonstrated similar changes in other MRT cell lines. Finally, reduced expression of SNF5 in normal human fibroblasts led to altered levels of these same complex members. These data establish that SNF5 loss during MRT development alters the repertoire of available SWI/SNF complexes, generally disrupting those associated with cellular differentiation. These findings support a model where SNF5 inactivation blocks the conversion of growth-promoting SWI/SNF complexes to differentiation-inducing ones. Therefore, restoration of these complexes in tumors cells provides an attractive approach for the treatment of MRTs.
Implications: SNF5 loss dramatically alters SWI/SNF complex composition and prevents formation of complexes required for cellular differentiation.
©2014 American Association for Cancer Research.
Conflict of interest statement
The authors declare that they have no conflicts of interest relating to these studies.
Figures




Similar articles
-
SNF5 reexpression in malignant rhabdoid tumors regulates transcription of target genes by recruitment of SWI/SNF complexes and RNAPII to the transcription start site of their promoters.Mol Cancer Res. 2013 Mar;11(3):251-60. doi: 10.1158/1541-7786.MCR-12-0390. Epub 2013 Jan 30. Mol Cancer Res. 2013. PMID: 23364536 Free PMC article.
-
The requirement for SNF5/INI1 in adipocyte differentiation highlights new features of malignant rhabdoid tumors.Oncogene. 2008 Mar 27;27(14):2035-44. doi: 10.1038/sj.onc.1210847. Epub 2007 Oct 8. Oncogene. 2008. PMID: 17922027
-
Fibroblast growth factor receptors as novel therapeutic targets in SNF5-deleted malignant rhabdoid tumors.PLoS One. 2013 Oct 30;8(10):e77652. doi: 10.1371/journal.pone.0077652. eCollection 2013. PLoS One. 2013. PMID: 24204904 Free PMC article.
-
Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth.Cancer Genet. 2014 Sep;207(9):365-72. doi: 10.1016/j.cancergen.2014.04.004. Epub 2014 Apr 13. Cancer Genet. 2014. PMID: 24853101 Free PMC article. Review.
-
Recent insights into the SWI/SNF complex and the molecular mechanism of hSNF5 deficiency in rhabdoid tumors.Cancer Med. 2023 Aug;12(15):16323-16336. doi: 10.1002/cam4.6255. Epub 2023 Jun 14. Cancer Med. 2023. PMID: 37317642 Free PMC article. Review.
Cited by
-
Lysine Methylation-Dependent Proteolysis by the Malignant Brain Tumor (MBT) Domain Proteins.Int J Mol Sci. 2024 Feb 13;25(4):2248. doi: 10.3390/ijms25042248. Int J Mol Sci. 2024. PMID: 38396925 Free PMC article. Review.
-
Comprehensive Molecular Characterization Identifies Distinct Genomic and Immune Hallmarks of Renal Medullary Carcinoma.Cancer Cell. 2020 May 11;37(5):720-734.e13. doi: 10.1016/j.ccell.2020.04.002. Epub 2020 Apr 30. Cancer Cell. 2020. PMID: 32359397 Free PMC article.
-
Chromatin remodeling and cancer: the critical influence of the SWI/SNF complex.Epigenetics Chromatin. 2025 Apr 23;18(1):22. doi: 10.1186/s13072-025-00590-w. Epigenetics Chromatin. 2025. PMID: 40269969 Free PMC article. Review.
-
Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers.Nat Struct Mol Biol. 2018 Jan;25(1):61-72. doi: 10.1038/s41594-017-0007-3. Epub 2017 Dec 11. Nat Struct Mol Biol. 2018. PMID: 29323272 Free PMC article.
-
Remodeling the cancer epigenome: mutations in the SWI/SNF complex offer new therapeutic opportunities.Expert Rev Anticancer Ther. 2019 May;19(5):375-391. doi: 10.1080/14737140.2019.1605905. Epub 2019 May 13. Expert Rev Anticancer Ther. 2019. PMID: 30986130 Free PMC article. Review.
References
-
- Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4:133–42. - PubMed
-
- Biswas A, Goyal S, Puri T, Das P, Sarkar C, Julka PK, et al. Atypical teratoid rhabdoid tumor of the brain: case series and review of literature. Child’s nervous system: ChNS: official journal of the International Society for Pediatric Neurosurgery. 2009;25:1495–500. - PubMed
-
- Wei D, Weissman BE. eLS. Vol. 2014 John Wiley & Sons Ltd; Chichester: 2014. Genetics and Genomics of Malignant Rhabdoid Tumors.
-
- Doan DN, Veal TM, Yan Z, Wang W, Jones SN, Imbalzano AN. Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene. 2004;23:3462–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

