LRRK2 kinase activity and biology are not uniformly predicted by its autophosphorylation and cellular phosphorylation site status
- PMID: 25009464
- PMCID: PMC4068021
- DOI: 10.3389/fnmol.2014.00054
LRRK2 kinase activity and biology are not uniformly predicted by its autophosphorylation and cellular phosphorylation site status
Abstract
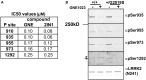
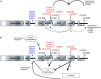
Missense mutations in the Leucine-Rich Repeat protein Kinase 2 (LRRK2) gene are the most common genetic predisposition to develop Parkinson's disease (PD) (Farrer et al., 2005; Skipper et al., 2005; Di Fonzo et al., 2006; Healy et al., 2008; Paisan-Ruiz et al., 2008; Lesage et al., 2010). LRRK2 is a large multi-domain phosphoprotein with a GTPase domain and a serine/threonine protein kinase domain whose activity is implicated in neuronal toxicity; however the precise mechanism is unknown. LRRK2 autophosphorylates on several serine/threonine residues across the enzyme and is found constitutively phosphorylated on Ser910, Ser935, Ser955, and Ser973, which are proposed to be regulated by upstream kinases. Here we investigate the phosphoregulation at these sites by analyzing the effects of disease-associated mutations Arg1441Cys, Arg1441Gly, Ala1442Pro, Tyr1699Cys, Ile2012Thr, Gly2019Ser, and Ile2020Thr. We also studied alanine substitutions of phosphosite serines 910, 935, 955, and 973 and specific LRRK2 inhibition on autophosphorylation of LRRK2 Ser1292, Thr1491, Thr2483 and phosphorylation at the cellular sites. We found that mutants in the Roc-COR domains, including Arg1441Cys, Arg1441His, Ala1442Pro, and Tyr1699Cys, can positively enhance LRRK2 kinase activity, while concomitantly inducing the dephosphorylation of the cellular sites. Mutation of the cellular sites individually did not affect LRRK2 intrinsic kinase activity; however, Ser910/935/955/973Ala mutations trended toward increased kinase activity of LRRK2. Increased cAMP levels did not lead to increased LRRK2 cellular site phosphorylation, 14-3-3 binding or kinase activity. In cells, inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser1292 by Calyculin A and Okadaic acid sensitive phosphatases, while the cellular sites are dephosphorylated by Calyculin A sensitive phosphatases. These findings indicate that comparative analysis of both Ser1292 and Ser910/935/955/973 phosphorylation sites will provide important and distinct measures of LRRK2 kinase and biological activity in vitro and in vivo.
Keywords: GTPase; LRRK2; Parkinson’s disease; kinase; kinase inhibitor; phosphorylation.
Figures




Similar articles
-
Lack of correlation between the kinase activity of LRRK2 harboring kinase-modifying mutations and its phosphorylation at Ser910, 935, and Ser955.PLoS One. 2014 May 16;9(5):e97988. doi: 10.1371/journal.pone.0097988. eCollection 2014. PLoS One. 2014. PMID: 24836358 Free PMC article.
-
Phosphorylation of LRRK2 serines 955 and 973 is disrupted by Parkinson's disease mutations and LRRK2 pharmacological inhibition.J Neurochem. 2012 Jan;120(1):37-45. doi: 10.1111/j.1471-4159.2011.07537.x. Epub 2011 Nov 11. J Neurochem. 2012. PMID: 22004453
-
Screening for novel LRRK2 inhibitors using a high-throughput TR-FRET cellular assay for LRRK2 Ser935 phosphorylation.PLoS One. 2012;7(8):e43580. doi: 10.1371/journal.pone.0043580. Epub 2012 Aug 28. PLoS One. 2012. PMID: 22952710 Free PMC article.
-
Pharmacological inhibition of LRRK2 cellular phosphorylation sites provides insight into LRRK2 biology.Biochem Soc Trans. 2012 Oct;40(5):1158-62. doi: 10.1042/BST20120137. Biochem Soc Trans. 2012. PMID: 22988882 Review.
-
LRRK2 Phosphorylation.Adv Neurobiol. 2017;14:51-70. doi: 10.1007/978-3-319-49969-7_3. Adv Neurobiol. 2017. PMID: 28353278 Review.
Cited by
-
An Assessment of LRRK2 Serine 935 Phosphorylation in Human Peripheral Blood Mononuclear Cells in Idiopathic Parkinson's Disease and G2019S LRRK2 Cohorts.J Parkinsons Dis. 2020;10(2):623-629. doi: 10.3233/JPD-191786. J Parkinsons Dis. 2020. PMID: 32007961 Free PMC article.
-
Genetic Modifiers of Neurodegeneration in a Drosophila Model of Parkinson's Disease.Genetics. 2018 Aug;209(4):1345-1356. doi: 10.1534/genetics.118.301119. Epub 2018 Jun 15. Genetics. 2018. PMID: 29907646 Free PMC article.
-
Abrogation of LRRK2 dependent Rab10 phosphorylation with TLR4 activation and alterations in evoked cytokine release in immune cells.Neurochem Int. 2021 Jul;147:105070. doi: 10.1016/j.neuint.2021.105070. Epub 2021 May 15. Neurochem Int. 2021. PMID: 34004238 Free PMC article.
-
Functional and behavioral consequences of Parkinson's disease-associated LRRK2-G2019S mutation.Biochem Soc Trans. 2018 Dec 17;46(6):1697-1705. doi: 10.1042/BST20180468. Epub 2018 Dec 4. Biochem Soc Trans. 2018. PMID: 30514770 Free PMC article. Review.
-
LRRK 2 gene mutations in the pathophysiology of the ROCO domain and therapeutic targets for Parkinson's disease: a review.J Biomed Sci. 2018 Jun 14;25(1):52. doi: 10.1186/s12929-018-0454-0. J Biomed Sci. 2018. PMID: 29903014 Free PMC article. Review.
References
-
- Alegre-Abarrategui J., Christian H., Lufino M. M., Mutihac R., Venda L. L., Ansorge O., et al. (2009). LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum. Mol. Genet. 18, 4022–4034 10.1093/hmg/ddp346 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

