Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon
- PMID: 25009735
- PMCID: PMC4089931
- DOI: 10.1186/2041-1480-5-21
Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon
Abstract
Background: Elucidating disease and developmental dysfunction requires understanding variation in phenotype. Single-species model organism anatomy ontologies (ssAOs) have been established to represent this variation. Multi-species anatomy ontologies (msAOs; vertebrate skeletal, vertebrate homologous, teleost, amphibian AOs) have been developed to represent 'natural' phenotypic variation across species. Our aim has been to integrate ssAOs and msAOs for various purposes, including establishing links between phenotypic variation and candidate genes.
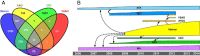
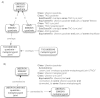
Results: Previously, msAOs contained a mixture of unique and overlapping content. This hampered integration and coordination due to the need to maintain cross-references or inter-ontology equivalence axioms to the ssAOs, or to perform large-scale obsolescence and modular import. Here we present the unification of anatomy ontologies into Uberon, a single ontology resource that enables interoperability among disparate data and research groups. As a consequence, independent development of TAO, VSAO, AAO, and vHOG has been discontinued.
Conclusions: The newly broadened Uberon ontology is a unified cross-taxon resource for metazoans (animals) that has been substantially expanded to include a broad diversity of vertebrate anatomical structures, permitting reasoning across anatomical variation in extinct and extant taxa. Uberon is a core resource that supports single- and cross-species queries for candidate genes using annotations for phenotypes from the systematics, biodiversity, medical, and model organism communities, while also providing entities for logical definitions in the Cell and Gene Ontologies. THE ONTOLOGY RELEASE FILES ASSOCIATED WITH THE ONTOLOGY MERGE DESCRIBED IN THIS MANUSCRIPT ARE AVAILABLE AT: http://purl.obolibrary.org/obo/uberon/releases/2013-02-21/ CURRENT ONTOLOGY RELEASE FILES ARE AVAILABLE ALWAYS AVAILABLE AT: http://purl.obolibrary.org/obo/uberon/releases/
Keywords: Bio-ontology; Evolutionary biology; Morphological variation; Phenotype; Semantic integration.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

