Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures
- PMID: 25013176
- PMCID: PMC4117792
- DOI: 10.1093/nar/gku579
Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures
Abstract
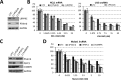
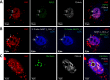
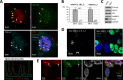
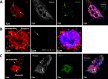
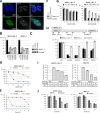
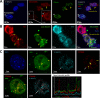
Nuclear paraspeckles are built co-transcriptionally around a long non-coding RNA, NEAT1. Here we report that transfected 20-mer phosphorothioate-modified (PS) antisense oligonucleotides (ASOs) can recruit paraspeckle proteins to form morphologically normal and apparently functional paraspeckle-like structures containing no NEAT1 RNA. PS-ASOs can associate with paraspeckle proteins, including P54nrb, PSF, PSPC1 and hnRNPK. NEAT1 RNA can be displaced by transfected PS-ASO from paraspeckles and rapidly degraded. Co-localization of PS-ASOs with P54nrb was observed in canonical NEAT1-containing paraspeckles, in perinucleolar caps upon transcriptional inhibition, and importantly, in paraspeckle-like or filament structures lacking NEAT1 RNA. The induced formation of paraspeckle-like and filament structures occurred in mouse embryonic stem cells expressing little or no NEAT1 RNA, suggesting that PS-ASOs can serve as seeding molecules to assemble paraspeckle-like foci in the absence of NEAT1 RNA. Moreover, CTN, an RNA reported to be functionally retained in paraspeckles, was also observed to localize to paraspeckle-like structures, implying that paraspeckle-like structures assembled on PS-ASOs are functional. Together, our results indicate that functional paraspeckles can form with short nucleic acids other than NEAT1 RNA.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Spector D.L. Nuclear domains. J. Cell Sci. 2001;114:2891–2893. - PubMed
-
- Shevtsov S.P., Dundr M. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2011;13:167–173. - PubMed
-
- Sone M., Hayashi T., Tarui H., Agata K., Takeichi M., Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007;120:2498–2506. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

