Ubiquitin signaling: extreme conservation as a source of diversity
- PMID: 25014160
- PMCID: PMC4197634
- DOI: 10.3390/cells3030690
Ubiquitin signaling: extreme conservation as a source of diversity
Abstract
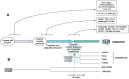
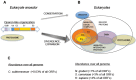
Around 2 × 103-2.5 × 103 million years ago, a unicellular organism with radically novel features, ancestor of all eukaryotes, dwelt the earth. This organism, commonly referred as the last eukaryotic common ancestor, contained in its proteome the same functionally capable ubiquitin molecule that all eukaryotic species contain today. The fact that ubiquitin protein has virtually not changed during all eukaryotic evolution contrasts with the high expansion of the ubiquitin system, constituted by hundreds of enzymes, ubiquitin-interacting proteins, protein complexes, and cofactors. Interestingly, the simplest genetic arrangement encoding a fully-equipped ubiquitin signaling system is constituted by five genes organized in an operon-like cluster, and is found in archaea. How did ubiquitin achieve the status of central element in eukaryotic physiology? We analyze here the features of the ubiquitin molecule and the network that it conforms, and propose notions to explain the complexity of the ubiquitin signaling system in eukaryotic cells.
Figures


References
-
- Ciechanover A., Elias S., Heller H., Ferber S., Hershko A. Characterization of the heat-stable polypeptide of the ATP-dependent proteolytic system from reticulocytes. J. Biol. Chem. 1980;255:7525–7528. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

