Lantibiotic immunity: inhibition of nisin mediated pore formation by NisI
- PMID: 25014359
- PMCID: PMC4094520
- DOI: 10.1371/journal.pone.0102246
Lantibiotic immunity: inhibition of nisin mediated pore formation by NisI
Abstract
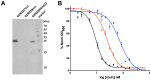
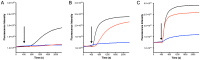
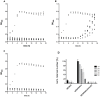
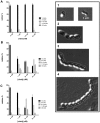
Nisin, a 3.4 kDa antimicrobial peptide produced by some Lactococcus lactis strains is the most prominent member of the lantibiotic family. Nisin can inhibit cell growth and penetrates the target Gram-positive bacterial membrane by binding to Lipid II, an essential cell wall synthesis precursor. The assembled nisin-Lipid II complex forms pores in the target membrane. To gain immunity against its own-produced nisin, Lactococcus lactis is expressing two immunity protein systems, NisI and NisFEG. Here, we show that the NisI expressing strain displays an IC50 of 73 ± 10 nM, an 8-10-fold increase when compared to the non-expressing sensitive strain. When the nisin concentration is raised above 70 nM, the cells expressing full-length NisI stop growing rather than being killed. NisI is inhibiting nisin mediated pore formation, even at nisin concentrations up to 1 µM. This effect is induced by the C-terminus of NisI that protects Lipid II. Its deletion showed pore formation again. The expression of NisI in combination with externally added nisin mediates an elongation of the chain length of the Lactococcus lactis cocci. While the sensitive strain cell-chains consist mainly of two cells, the NisI expressing cells display a length of up to 20 cells. Both results shed light on the immunity of lantibiotic producer strains, and their survival in high levels of their own lantibiotic in the habitat.
Conflict of interest statement
Figures
References
-
- Cleveland J, Montville TJ, Nes IF, Chikindas ML (2001) Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol 71: 1–20. - PubMed
-
- Cotter PD, Hill C, Ross RP (2005) Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3: 777–788. - PubMed
-
- Kruszewska D, Sahl HG, Bierbaum G, Pag U, Hynes SO, et al. (2004) Mersacidin eradicates methicillin-resistant Staphylococcus aureus (MRSA) in a mouse rhinitis model. J Antimicrob Chemother 54: 648–653. - PubMed
-
- Galvin M, Hill C, Ross RP (1999) Lacticin 3147 displays activity in buffer against gram-positive bacterial pathogens which appear insensitive in standard plate assays. Lett Appl Microbiol 28: 355–358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

