Virtual finger boosts three-dimensional imaging and microsurgery as well as terabyte volume image visualization and analysis
- PMID: 25014658
- PMCID: PMC4104457
- DOI: 10.1038/ncomms5342
Virtual finger boosts three-dimensional imaging and microsurgery as well as terabyte volume image visualization and analysis
Abstract
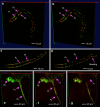
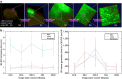
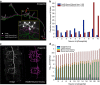
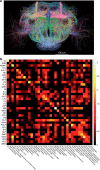
Three-dimensional (3D) bioimaging, visualization and data analysis are in strong need of powerful 3D exploration techniques. We develop virtual finger (VF) to generate 3D curves, points and regions-of-interest in the 3D space of a volumetric image with a single finger operation, such as a computer mouse stroke, or click or zoom from the 2D-projection plane of an image as visualized with a computer. VF provides efficient methods for acquisition, visualization and analysis of 3D images for roundworm, fruitfly, dragonfly, mouse, rat and human. Specifically, VF enables instant 3D optical zoom-in imaging, 3D free-form optical microsurgery, and 3D visualization and annotation of terabytes of whole-brain image volumes. VF also leads to orders of magnitude better efficiency of automated 3D reconstruction of neurons and similar biostructures over our previous systems. We use VF to generate from images of 1,107 Drosophila GAL4 lines a projectome of a Drosophila brain.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

