Genome-wide analysis of trans-splicing in the nematode Pristionchus pacificus unravels conserved gene functions for germline and dauer development in divergent operons
- PMID: 25015138
- PMCID: PMC4138322
- DOI: 10.1261/rna.041954.113
Genome-wide analysis of trans-splicing in the nematode Pristionchus pacificus unravels conserved gene functions for germline and dauer development in divergent operons
Abstract
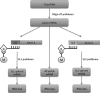
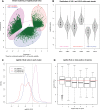
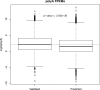
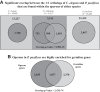
Discovery of trans-splicing in multiple metazoan lineages led to the identification of operon-like gene organization in diverse organisms, including trypanosomes, tunicates, and nematodes, but the functional significance of such operons is not completely understood. To see whether the content or organization of operons serves similar roles across species, we experimentally defined operons in the nematode model Pristionchus pacificus. We performed affinity capture experiments on mRNA pools to specifically enrich for transcripts that are trans-spliced to either the SL1- or SL2-spliced leader, using spliced leader-specific probes. We obtained distinct trans-splicing patterns from the analysis of three mRNA pools (total mRNA, SL1 and SL2 fraction) by RNA-seq. This information was combined with a genome-wide analysis of gene orientation and spacing. We could confirm 2219 operons by RNA-seq data out of 6709 candidate operons, which were predicted by sequence information alone. Our gene order comparison of the Caenorhabditis elegans and P. pacificus genomes shows major changes in operon organization in the two species. Notably, only 128 out of 1288 operons in C. elegans are conserved in P. pacificus. However, analysis of gene-expression profiles identified conserved functions such as an enrichment of germline-expressed genes and higher expression levels of operonic genes during recovery from dauer arrest in both species. These results provide support for the model that a necessity for increased transcriptional efficiency in the context of certain developmental processes could be a selective constraint for operon evolution in metazoans. Our method is generally applicable to other metazoans to see if similar functional constraints regulate gene organization into operons.
Keywords: gene order; genome evolution; operon; trans-splicing.
© 2014 Sinha et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
-
- Blaxter M, Liu L 1996. Nematode spliced leaders: ubiquity, evolution and utility. Int J Parasitol 26: 1025–1033 - PubMed
-
- Blumenthal T 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet 11: 132–136 - PubMed
-
- Blumenthal T, Gleason KS 2003. Caenorhabditis elegans operons: form and function. Nat Rev Genet 4: 112–120 - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- SRA/SRP039388
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
