SAS-6 assembly templated by the lumen of cartwheel-less centrioles precedes centriole duplication
- PMID: 25017693
- PMCID: PMC4116473
- DOI: 10.1016/j.devcel.2014.05.008
SAS-6 assembly templated by the lumen of cartwheel-less centrioles precedes centriole duplication
Erratum in
- Dev Cell. 2014 Aug 25;30(4):488
Abstract
Centrioles are 9-fold symmetric structures duplicating once per cell cycle. Duplication involves self-oligomerization of the centriolar protein SAS-6, but how the 9-fold symmetry is invariantly established remains unclear. Here, we found that SAS-6 assembly can be shaped by preexisting (or mother) centrioles. During S phase, SAS-6 molecules are first recruited to the proximal lumen of the mother centriole, adopting a cartwheel-like organization through interactions with the luminal wall, rather than via their self-oligomerization activity. The removal or release of luminal SAS-6 requires Plk4 and the cartwheel protein STIL. Abolishing either the recruitment or the removal of luminal SAS-6 hinders SAS-6 (or centriole) assembly at the outside wall of mother centrioles. After duplication, the lumen of engaged mother centrioles becomes inaccessible to SAS-6, correlating with a block for reduplication. These results lead to a proposed model that centrioles may duplicate via a template-based process to preserve their geometry and copy number.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

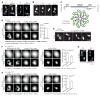


Comment in
-
Mother centrioles do a cartwheel to produce just one daughter.Dev Cell. 2014 Jul 28;30(2):111-2. doi: 10.1016/j.devcel.2014.07.013. Dev Cell. 2014. PMID: 25073149 Free PMC article.
References
-
- Arquint C, Nigg EA. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Current biology : CB. 2014;24:351–360. - PubMed
-
- Arquint C, Sonnen KF, Stierhof YD, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. Journal of cell science. 2012;125:1342–1352. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

