Substrate-bound outward-open state of the betaine transporter BetP provides insights into Na+ coupling
- PMID: 25023443
- PMCID: PMC4745115
- DOI: 10.1038/ncomms5231
Substrate-bound outward-open state of the betaine transporter BetP provides insights into Na+ coupling
Abstract
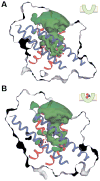
The Na(+)-coupled betaine symporter BetP shares a highly conserved fold with other sequence unrelated secondary transporters, for example, with neurotransmitter symporters. Recently, we obtained atomic structures of BetP in distinct conformational states, which elucidated parts of its alternating-access mechanism. Here, we report a structure of BetP in a new outward-open state in complex with an anomalous scattering substrate, adding a fundamental piece to an unprecedented set of structural snapshots for a secondary transporter. In combination with molecular dynamics simulations these structural data highlight important features of the sequential formation of the substrate and sodium-binding sites, in which coordinating water molecules play a crucial role. We observe a strictly interdependent binding of betaine and sodium ions during the coupling process. All three sites undergo progressive reshaping and dehydration during the alternating-access cycle, with the most optimal coordination of all substrates found in the closed state.
Figures


References
-
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 2005;437:215–223. - PubMed
-
- Perez C, Koshy C, Yildiz O, Ziegler C. Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature. 2012;490:126–130. - PubMed
-
- Forrest LR, Kramer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim Biophys Acta. 2011;1807:167–188. - PubMed
-
- Wong FH, et al. The Amino Acid-Polyamine-Organocation Superfamily. Journal of Molecular Microbiology and Biotechnology. 2012;22:105–113. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

